1 Introduction
Sustainable data management is fundamental in today’s data-driven world for several reasons. The amount of acquired data storage capacity has long ceased to be the limiting factor, while the computing power has increased greatly [1]. However, it is the ability to share data rather than generate it that defines success [2]. Furthermore, interdisciplinary and international collaborations have become essential in scientific research, and the main means of communication is based on digital documents [3]. A bottleneck in data exploration and processing, and therefore the general re-usability, is often the lack of auxiliary data, i.e. metadata. As a consequence, much time is spent on obtaining missing information. In some cases, this may require to re-conduct simulations and experiments. Effective data management practices hence hold the potential of saving time and money as well as increasing the value of data at the same time.
Introducing a new data management concept can be challenging due to conflicting priorities, expectations, and existing practices, as well as a lack of expertise or clear understanding of the benefits. Efficient use of standards is crucial for large and interdisciplinary collaborations. While those groups have developed domain-specific solutions, small research groups and PhD projects face challenges due to the use of multiple file formats, individual software solutions, personal preferences for storage and tools and established structures [4]. Common issues are the lack of time and resources to develop and implement a comprehensive and sustainable data management approach [5], which fulfills the requirements of the community and good scientific practice. Therefore, flexible and manageable solutions are needed to address this issue.
Although the implementation of a common management system is beneficial in the long term, both financially [6] and in terms of efficiency, it disrupts structures and requires time, resources and cultural change. In academia, high staff turnover is an additional barrier, making it difficult to establish sustainable solutions. The decay of value develops as projects progress, ultimately finish and contracts expire. Consequently, the value of data will diminish over time. This issue is discussed in more detail in [7], [8]. In addition, a value decay can also be observed with increasing distance from the source of the data. The further away and therefore less involved a potential data user is, the more information may be missing, either due to restricted access or limited personal connections. Ensuring that data is preserved and being interpretable at all times can be achieved by adhering to the so-called FAIR principles, which stand for Findable, Accessible, Interoperable and Reusable and were first introduced in 2016 by [9]. Since their publication, the principles have become the cornerstones of many scientific communities and help to establish a sustainable data management [10]. Structured, highly descriptive information about data, known as metadata, is an integral part of it. Metadata provides context about its creation, purpose, use, processing history and the meaning of datasets. Consequently, it enables data to be discoverable, interoperable and reusable.
This work is a contribution to assist small collaborative groups or communities and doctoral researchers with achieving a FAIR research data lifecycle by using the HDF5 (Hierarchical Data Format) file format. These groups are often faced with challenges such as heterogeneous file formats, the absence of standards within their fields, and limited expertise and resources for sustainable data management. The paper describes the scope and concepts of a Python package named h5rdmtoolbox and how it facilitates the implementation of FAIR principles using the HDF5 file format. Complementing this manuscript, an extensive online documentation is provided [11], leveraging Jupyter Notebooks [12]. This documentation offers in-depth insights and additional examples for immediate usage, serving as a comprehensive resource for users seeking detailed information and practical guidance.
1.1 Outline of the paper
Firstly, the paper outlines the package’s scope in comparison to existing and related works. This is followed by a section stating the concepts and architecture of the toolbox, describing the applied design principles and methods. Subsequently, the paper discusses concrete implementation details of all sub-packages and provides illustrative examples, referencing to their relevance within the research data lifecycle. Limitations of the presented package are stated before the paper concludes and summarizes the presented work. An outlook is given on future developments and potential enhancements.
2 Scope and related work
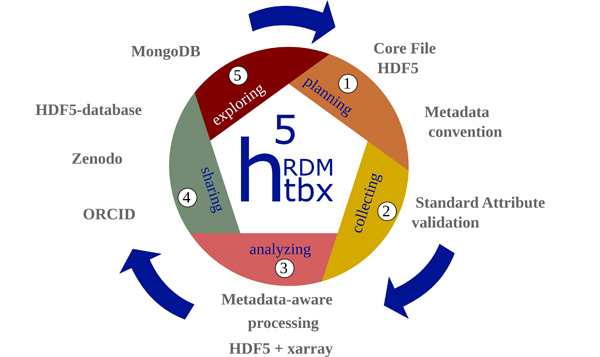
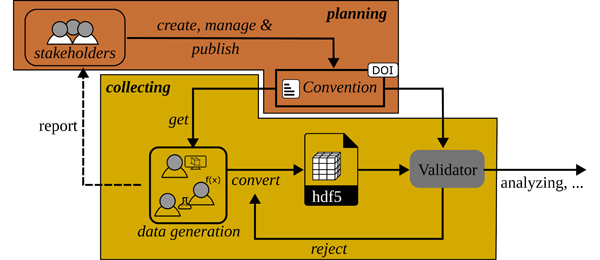
The primary aim of this toolbox is to offer comprehensive support throughout the lifecycle of research data (c.f. Figure 1) for small collaborative groups, communities, and doctoral researchers engaged in utilizing or contemplating the use of HDF5 files as their central file format. The file format is selected for various reasons, which are stated hereafter. A review of other file formats is beyond the scope of this work and literature should be referred to, for example [2], [8], [13].
Figure 1: Illustration of the lifecycle of research data. Each phase is supported by the h5rdmtoolbox. It starts by selecting a file format (HDF5) and a metadata concept (1) and performing quality assurance measures during the selection and processing phase (2). Data is analyzed effectively for scientific output in the next step (3). After publication, the availability of the data should be ensured (4). (Meta)data quality finally is defined by its findability and consequently its re-usability (5) for additional analysis at later time. The respective tools and solutions provided by the toolbox are indicated by keywords around the lifecycle and explained in this work.
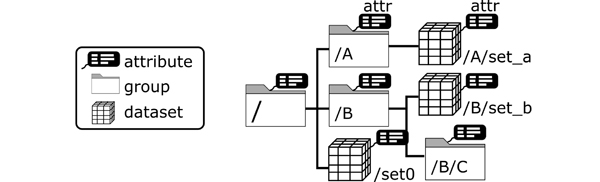
HDF5 features efficient storing of large multidimensional datasets together with metadata independent of the storage media, programming environment or operating system. The hierarchical structure of group and dataset objects (cf. Figure 2) resembles most engineering data. Attributes (key-value pairs) are means to store metadata and can be assigned to each object. The HDF5 file format is therefore regarded as self-explanatory. HDF5 finds application in numerous scientific domains, such as earth observation [14], high-energy physics [15] or fluid dynamics [16]. An in-depth presentation of the file format can be found in [17].
Figure 2: Illustration of the hierarchical structure of an HDF file. The internal file structure is organized like a file storage system, where folders are represented by the HDF group objects and files by HDF dataset objects. Both objects can be associated with attributes, which provide the metadata in order to make the objects interpretable.
Despite all the advantages of the file format, the organization of data management around HDF5 is left to the user [18]. This means that the choice of attribute names and values is not regulated by any standard. Findability, effective re-usability and automatic analysis, however, are dependent on standardization [19].
The necessity for designing management solutions around the HDF5 file format is therefore evident. While existing solutions, such as proposed in [8], [14], [15], [16], [20], [21] address this need, they are often domain-specific, primarily focused on efficiently meeting the demands of specific communities rather than providing a generalized framework applicable to diverse problems. For example, formats like Nexus [15] or Photon-HDF5 [21] prescribe specific group and dataset organizations and metadata usage tailored to their respective data sources, such as neutron and X-ray data and molecule spectroscopy experiments, respectively. Other libraries like Zarr [20] address challenges associated with very large data (terabyte-scale) in the field of bioimaging with a particular emphasis on optimized cloud-based operations and the sharing of HDF5-based datasets. Finally, the issue of efficient database solutions for HDF5 are addressed in [1], [22].
Besides the specificity of the solutions, adopting aforementioned solutions to new problems is very difficult due to their complexity and required expertise in the field. When data management solutions are needed for a concrete projects, it is crucial to minimize entry barriers. Currently, for HDF5, a general approach to manage data in all aspects during its lifecycle including metadata concepts, database solutions and practical interfaces are missing. The presented Python package h5rdmtoolbox seeks to bridge the gap between the advanced communities with domain-specific solutions and researchers trying to manage their data without established standards in place. Leveraging well-established Python packages, this toolbox offers high-level tools and interfaces within one package, that actively contribute to the promotion of FAIR data creation. As a whole, the package seeks to be a central resource of tools for scientists allowing them to manage their HDF5 data along the full data lifecycle from planning (1) via acquisition (2) and analysis to publication in data repositories (4) and sharing in databases (5). Figure 1 illustrates these stages and relates keywords to features of the toolbox.
3 Concepts and architecture of the toolbox
A key aspect of the toolbox lies in its adaptable implementation of metadata standards and interfaces to databases and repositories, allowing it to be relevant across many research fields with varying requirements. The challenge is to attain this flexibility without introducing excessive complexity, all while ensuring adherence to the FAIR principles. The toolbox achieves this through four principles:
Relevant programming language: The choice of programming language significantly impacts the usability and acceptance of this toolbox, as well as data handling in general. Python is selected for this purpose due to its status as one of the most popular and widely used language in the scientific community. The high relevance of Python in the field allows the toolbox to address as many users as possible.
One core file format: The Hierarchical Data Format (HDF5) [23] is selected as the core and general purpose file format. It is suitable for most scientific and engineering data sources and allows metadata to be stored with the raw data, making it a self-explanatory data store. The file format is open-source, well-supported by the HDF Group [23] and has a proven track record in many disciplines. Opting for a single file format as the foundation for a management toolbox is, therefore, not limiting. Prioritizing user-friendliness and widespread acceptance, the toolbox implements high-level interfaces to HDF5, extending the capabilities of the commonly used Python package h5py [24].
Flexible Metadata Standardization: Enabling the storage of metadata alongside raw data necessitates its standardization (convention) to achieve discoverability. The toolbox introduces a simple and flexible definition of so-called standard attributes. Users can design their own convention, which provides feedback about the correctness of the (meta)data created. This ensures that users maintain the accuracy and completeness of their data and metadata.
Extensibility: Adaptability extends beyond just metadata standards; it encompasses various aspects of the toolbox, including interfaces to databases and data repositories. Abstract classes establish communication rules between HDF5 and users, enabling the community to add new interfaces on top of the currently implemented ones and to make them available to others through the toolbox.
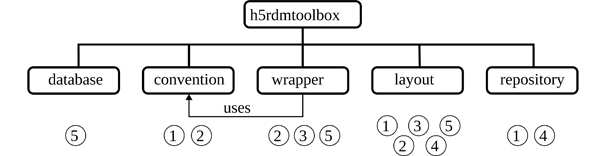
In this work, a five-stage representation of the research data lifecycle is adopted, as illustrated in Figure 1. This framework forms the basis for the toolbox’s architectural design, aligning its functionalities with the key stages of the data lifecycle. Consequently, the toolbox is structured into five sub-packages, as depicted in Figure 3. The numerical assignments in the figure directly correlate with the roles of these sub-packages in the stages of the data lifecycle (c.f. Figure 1). This structured approach enhances the toolbox’s utility by providing specialized tools for each phase of the research data lifecycle.
Figure 3: Organization of the sub-packages in the presented package. The core module is called wrapper, which adds useful functionality for the user when interacting with HDF5 files. It uses the convention module to manage metadata requirements when creating and reading data. The other modules are not dependent on each other and must be imported on demand. The numbers indicate their main areas of application within the different stages of the data lifecycle, as shown in Figure 1.
The components of the sub-packages are designed in a manner that ensures independence from each other, facilitating individual development and modularity. One exception is made to the sub-packages wrapper and convention. The following sections will highlight the features and implementations of the sub-packages, as well as their importance within the data lifecycle.
3.1 layout
Research projects start with a scientific question and a data management plan (DMP) [25]. The DMP outlines how data is handled during and after the project. One important aspect is the agreement on common exchange formats (in this work HDF5). It has a significant impact on the realization of a FAIR data cycle as a whole, especially, when it comes to sharing data [20]. Besides a common vocabulary, the internal structure (layout) of the file is important. It is the basis for reliable processing and automated analysis. The hierarchical structure of HDF5 files allows various strategies to organize data and therefore must be regulated by the project data manager.
The sub-package layout implements the class Layout, which is a collection of specifications. Each specification is a query that is executed during the validation of a file. The rationale behind this approach is that all elements, that are expected to be present in a file, must be identifiable. The code in Listing 1 illustrates the definition of a Layout: First, the layout object is created. Then two specifications are added by providing a query function and query parameters (using pseudocode for simplicity). The initial parameter may be any Python function that is capable of accepting the query parameters and returning a list of identified HDF5 objects. As part of the toolbox and its documentation [11], a database solution is provided within the sub-package database.
1 from h5rdmtoolbox import layout
2 lay = layout.Layout()
3 spec1 = lay.add(func=query_function,
4 query=<has dataset with name 'x_velocity'>)
5 spec2 = lay.add(func=query_function,
6 query=<datasets have the attribute 'units'>)
7 lay.validate("filename.hdf")
Listing 1: Code example for defining a Layout to validate HDF5 files based on query statements. The queries are written in pseudocode for enhanced readability.
The Layout concept should be part of every phase in the data lifecycle. Once the definition has been established during the planning phase (1), it is advisable to validate the integrity of the file at each stage. This is because the content may have been altered in the meantime, for example, the agreed internal setup or used attributes may have been modified. Verifying that the layout remains consistent with the intended definition is essential for the generation of reliable data and complete files. Avoiding missing information through careful definition of the file content in combination with regular checks is the basis of FAIR data.
3.2 convention
In addition to a robust HDF5 layout, the provision of meaningful and comprehensive metadata for HDF5 datasets and groups is of the utmost importance. This ensures that files are interpretable by both humans and machines. During the planning phase (1), a selection of relevant attributes for the investigated problem is important. The quality of these attributes significantly influences the findability of data within an HDF5 file, as well as the reusability and interoperability aspects of FAIR data in general. The term ”quality” here refers to whether attributes are linked to existing metadata concepts that can be referenced to persistent sources. Examples include controlled vocabularies such as the CF Convention [19], metadata schemas like EngMeta [26], or ontologies like Metadata4ing [27]. These sources provide standardized and well-defined terms that enhance the clarity and consistency of metadata, contributing to improved data discoverability and reuse. Documentation for the toolbox [11] includes examples showcasing the possible utilization of these standards within the toolbox. The concept of Conventions is explained in the following.
The h5rdmtoolbox implements the concept of so-called standard attributes as part of a Convention object to validate relevant metadata, i.e. HDF5 attributes, during runtime as the user writes data to the file. The implementation is based on the Python package pydantic and hence reuses successful existing solutions. It should be noted, that this approach of attribute validation partly overlaps with the concept of Layouts. However, layout checks are performed after the file has been written and therefore allows for more complex requirements, that have been defined by stakeholders (e.g. dependency checks in the form of ”if a dataset is named X then it should be 1D and of data type float32”). The strength of using a Convention is, that it allows checks during data creation with immediate feedback. The focus is on usage of specific attributes and their correct usage. It is therefore especially helpful during software development, data manipulation and conversion.
Figure 4 illustrates a common workflow, which makes use of this concept. The stakeholders of a project define and share a set of standardized attributes of type StandardAttribute within a Convention. The latter is saved in a YAML file and is shared across all users, which are directly working with HDF5 files. By integrating the Convention into their workflows through the h5rdmtoolbox, they obtain direct feedback through a validation mechanism. As a result, the quality in terms of reliable and comprehensive data description through attributes is ensured and basis for the FAIRness of HDF5 file is set.
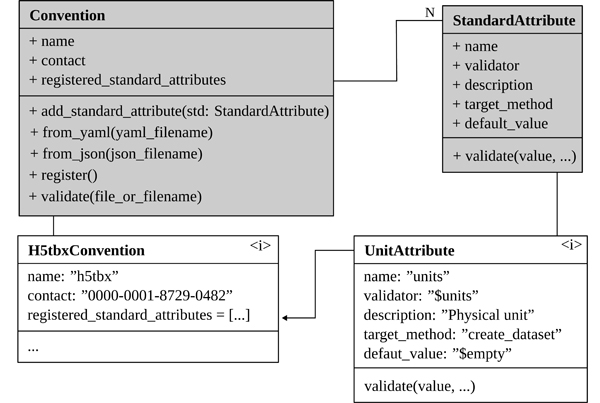
As shown in the class diagram in Figure 5, a Convention object takes a list of StandardAttribute objects. The important properties of a StandardAttribute are validator and target_method. The target_method assigns the object to a method of the h5py package (other options are __init__ or create_group) and the validator defines how the attribute is validated during assignment.
1 import h5rdmtoolbox as h5tbx
2 h5tbx.use("h5tbx")
3 with h5tbx.File(myfile.hdf, "r+") as h5:
4 h5.create_dataset("ds", data=4, units="m/s")
Listing 2: Minimal example of using a Convention. By enabling the ”h5tbx”, the standard attribute ”units” becomes obligatory in the method create_dataset. The value of ”units” is validated and automatically added to the newly created dataset or an error is raised.
A minimal example of the two instances shown in Figure 5 is written in code in Listing 2. Note, that the parameter ”units” in the function call is not part of the underlying h5py package but gets dynamically added by enabling the Convention. It is also noteworthy that by setting the keyword ”$empty” as the default value, the attribute becomes obligatory. For HDF5 datasets, this is in fact a reasonable choice, as generally physical data is written to datasets, which require a physical unit.
As indicated in the class diagram, Conventions can also be defined in files (JSON or YAML), which allows sharing the Convention via data repositories or databases with all involved stakeholders. By enabling the project Convention during file manipulating, users receive immediate feedback on the validity of the used standardized attributes (c.f. Figure 4). This is a difference to the concept of Layouts, which are static validators. For further information and examples about the implementation details, pre-implemented validators as well as the user-defined creation of new ones, please refer to the documentation [11], as this information exceeds the scope of this paper.
The documentation provides extensive details, practical examples, and guidance to support users in utilizing and customizing Conventions and validators within the h5rdmtoolbox.
3.3 wrapper
The package wrapper plays a central role within the toolbox by implementing a thin layer around the HDF5 Python library h5py. Besides user-friendly high-level methods for interactive representation of the file content in Jupyter Notebooks or helper methods for special attributes or datasets, the wrapper package is responsible for
integration of the Convention concept into the h5py framework and
metadata-aware exchange of data through xarray object [28].
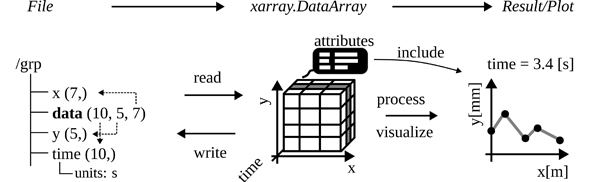
The integration of the xarray package into the toolbox provides several advantages. As previously highlighted, one of the reasons for selecting HDF5 is its compatibility with the multidimensionality of many scientific and engineering datasets, allowing the storage of attributes alongside the data. However, using numpy arrays as part of the h5py package results in the loss of two important sets of information. Firstly, numpy arrays can only represent array data, discarding attributes associated with the data. Secondly, the axis of a multidimensional array can only be addressed by their indices (0, 1, etc.), potentially losing references to other datasets in the HDF5 format (a concept known as dimension scales in h5py [24]). This limitation hinders the interpretation of values and their context.
The xarray package addresses these limitations by wrapping its functionality around numpy arrays [28]. It enables the association of attributes to the values and allows the labeling of the axes in multidimensional arrays. This structure closely aligns with the HDF5 dataset model. By returning ”metadata-aware” xarray objects, the toolbox ensures that provenance information is added, enhancing the intuitiveness and reliability of data processing. The auxiliary information is consistently preserved during data utilization for visualization or other post-processing steps, as depicted in Figure 6. It is noteworthy, that xarray has a strong plotting utility, that automatically extracts information from the data object, incorporating it into the labels and title of the plot. The synergies between HDF5 and xarray, resulting in benefits like concise code and interactive visualization of metadata, are best illustrated through practical examples. To gain a deeper understanding and explore enhanced workflows and data operations, it is recommended to consult the online documentation of the h5rdmtoolbox [11]. For the sake of completeness, a short example is given in the following.
The code example in Listing 3 demonstrates the workflow as illustrated in Figure 6. A subset of the dataset ”data” is selected based on the coordinates. The return value is a xarray.DataArray on which the rolling mean is computed. The result is finally plotted on the screen. With only a few lines of code, the user obtains quick insight into the dataset while maintaining comprehensibility and traceability. Another notable feature wrapped around the core h5py package is the ability to encode the semantics of HDF5 data using the concept of Resource Description Framework (RDF) [29]. Similar to the attribute manager interface (attrs) of h5py, the toolbox uses an RDF manager (rdf). It enables the user to enrich HDF5 attributes, datasets and groups with formal metadata using semantic RDF triples (subject - predicate - object). More information on the technology can be found in [30].
Listing 4 outlines a minimal example how metadata of a person can be precisely described using Internationalized Resource Identifiers (IRI). While the choice of dataset, group and attribute names is often based on personal preferences, RDF triples add meaning to the group ”contact” and its attributes. The assignment of globally unique and persistent identifiers plays a pivotal role in the fulfillment of the FAIR principles. Like this, metadata becomes interpretable by both humans and machines. The method dump_jsonld extracts the semantic information, which may be saved in a JSON-LD file or stored in a database for further usage. The internal HDF5 structure can also be described using RDF, but is disabled here (structural=False) to obtain a compact output. The documentation may be consulted for more details at this point.
1 import h5rdmtoolbox as h5tbx
2 with h5tbx.File(filename) as h5:
3 # select and read selected data and store in variable:
4 d = h5["data"].sel(x=4.3, y=0.2, method="nearest")
5
6 # process (compute rolling mean over time with window size 3):
7 drm = d.rolling(time=3).mean()
8
9 # visualize the result:
10 drm.plot()
Listing 3: Example of data extraction using the toolbox. The returned value is an xarray.DataArray containing comprehensive metadata from the underlying HDF5 dataset. This facilitates transparent data operations and minimizes potential errors. Additionaly, many operations can be reduced to one line of code, which makes scripts concise and traceable.
3.4 repository
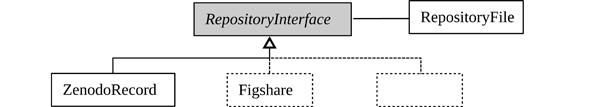
How data is shared depends on the scope and restrictions of the project (phase 4 in the lifecycle). Most use cases will, at least for some time, store data locally for internal use and later upload it to a data repository. The sub-package repository implements an abstract interface class to data repositories and their files. At the time of writing, one concrete realization of such an interface is implemented for Zenodo [33]. It is one of the most popular repositories in the scientific community to publish scientific data with open-access. Interfaces to other platforms are planned to be added in the future, such as Figshare [34] for example. The design of the repository sub-package explicitly promotes this by using an object-oriented design: An abstract base class RepositoryInterface defines mandatory properties and methods for the user-platform interaction, as depicted in Figure 7. Moreover, the interaction of users with files within a repository is prescribed by RepositoryFile. The chosen design streamlines and simplifies the data exchange with repositories (see Listing 5).
Figure 7: The architecture of the repository sub-package implements the basic interface (abstract class RepositoryInterface) to data repositories. Through One concrete implementation is provided for Zenodo (ZenodoRecord). The interface to files within the repository is realized through RepositoryFile, clearly defining common properties and download functionalities across all repositories. Other popular platforms (e.g. Figshare) could be added by code contributors to the h5rdmtoolbox. The gray components are abstract classes, white boxes indicate concrete implementations, and the dashed lines indicate potential extensions in future.
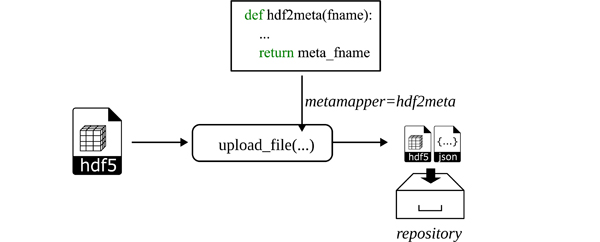
The repository interface class implements the method upload_file, which allows to automatically map metadata to a secondary file, which is uploaded alongside the original file (see Figure 8). This has the following reasoning: Large files are expensive to download in terms of time, especially if it turns out, that the data is not matching the expectations of a user. As data repositories typically only offer descriptive information regarding the type of data publication (e.g., creator, version, time, keywords, license, etc.), the content of large files can only be examined after they have been downloaded.
Figure 8: The workflow of uploading data to a repository involves a so-called metamapper function, which extracts metadata from and about the file (attributes and structure) and writes it into a JSON-LD file. This is done automatically for HDF5 files, unless the user provides a custom function (here representatively indicated with hdf2meta, resulting in an additional JSON file). Both files are uploaded to the repository. This procedure is especially helpful for large HDF5 files. Interested users may first download the metadata file and inspect the content before downloading the large file.
1 import h5rdmtoolbox as h5tbx
2
3 with h5tbx.File() as h5:
4 g = h5.create_group("contact")
5 g.attrs["name"] = "Probst"
6 g.attrs["oid"] = "0000-0001-8729-0482"
7
8 # enrich with RDF metadata:
9 g.rdf.type = "http://xmlns.com/foaf/0.1/Person"
10 g.rdf.subject = "https://orcid.org/0000-0001-8729-0482"
11 g.rdf.predicate["name"] = "http://xmlns.com/foaf/0.1/lastName"
12 g.rdf.predicate["oid"] = "http://w3id.org/nfdi4ing/metadata4ing#
13 orcidId"
14
15 print(h5.dump_jsonld(structural=False, indent=2, resolve_keys=True))
16
17 # output:
18 # {
19 # "@context": {
20 # "foaf": "http://xmlns.com/foaf/0.1/",
21 # "m4i": "http://w3id.org/nfdi4ing/metadata4ing#"
22 # },
23 # "@id": "https://orcid.org/0000-0001-8729-0482",
24 # "@type": "foaf:Person",
25 # "foaf:lastName": "Probst",
26 # "m4i:orcidId": "0000-0001-8729-0482"
27 # }
Listing 4: Simple example code highlighting the semantic enrichment of HDF5 data using RDF triples and globally unique identifiers from existing ontologies, here foaf [31] or m4i [32]. This approach ensures that attributes and groups are assigned a concise and clear meaning, which can be interpreted by machines and is therefore independent of the author’s or project’s context
Especially large HDF5 files may contain much and complex information, not only based on attributes but also from the internal structure and dataset properties. The automatic extraction of metadata is implemented for HDF5 files only as part of the toolbox and uses RDF as a universal metadata description (see e.g. Listing 4). If the user wish to use custom mappings for HDF5 files or other file formats before their upload, the custom function should be passed to the argument metamapper. In the example shown in Listing 5, an HDF5 file is uploaded using the built-in function hdf2jsonld from the wrapper. It writes the metadata into a JSON file using the JSON-LD format, resulting in a small text file. Another user exploring the repository may download the JSON file first, which allows investigating the HDF5 metadata content, and then eventually download the potentially large HDF5 file.
1 from h5rdmtoolbox.repository import zenodo
2 from h5rdmtoolbox.wrapper import hdf2jsonld
3
4 repo = zenodo.ZenodoRecord(None, sandbox=True) # new testing deposit
5 repo.upload_file("my_file.hdf", metamapper=hdf2jsonld, skipND=1)
6
7 meta_filename = repo.files["my_file.jsonld"].download()
8 # … review JSON-LD file and eventually download the HDF5 file
Listing 5: Example code demonstrating the upload process of HDF5 files. The metamapper parameter expects a function, which extracts metadata information from the HDF5 file and uploads it alongside the HDF5 file. The default function as used in the example uses hdf2json, which is a built-in function. It extracts the structure in the json-ld format. The parameter skipND is specific to hdf2json and is automatically passed to it.
3.5 database
Exploring HDF5 data and hence an efficient re-use requires a query mechanism for the files. The toolbox implements two ways:
Using HDF5 as a database inside a file system.
Mapping HDF5 to the NoSQL database MongoDB [35].
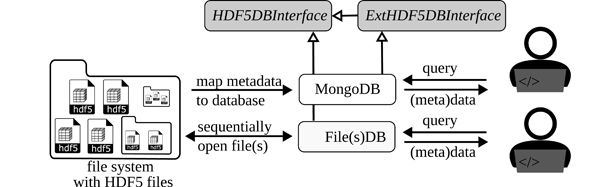
Figure 9 shows the workflows for both options. The simplest solution uses an HDF5 file itself as a database and multiple files as multiple databases respectively. For the case of one file the user calls the database interface class FileDB, for multiple files FilesDB. A query call is constructed similar to the one using a dedicated database solution, which is MongoDB. Each search will recursively walk through one or multiple HDF5 files. Yet simple, this approach may be inefficient for many or large HDF5 files.
Figure 9: Workflow of the two provided database solutions: The simplest solution using FileDB or FilesDB allows the user to perform queries directly on one or many HDF5 files, respectively. The requests are sequentially executed and scan the complete files. A more efficient solution maps the metadata to a NoSQL database (MongoDB). Here, query requests can be more complex and are more efficient but requires a prior mapping process, which the toolbox provides. Both implementations are inherited from HDF5DBInterface ensuring a common interface between user and database solution.
A second and more performant solution maps the attributes to a MongoDB database. A query on MongoDB is very efficient and allows more complex queries as compared to the current implementation of FileDB and FilesDB. Depending on the amount of files and their size, the extraction of metadata and writing to the database may be time-consuming. However, frequent query calls are processed very quickly, resulting in a faster overall solution. It should be noted, that MongoDB is used as a metadata database, which requires keeping the original HDF5 files. If no further queries are planned, the database can be deleted again.
Both approaches are implemented in the toolbox based on the abstract class HDF5DBInterface. This class defines two query methods (find and find_one). For implementations using external databases like MongoDB, inserting methods are required as defined in ExtHDF5DBInterface. The utilization of these classes defines the interface between the database and the users, and also serves as a foundation for future implementations of other third-party databases. The used syntax for the queries, the capabilities of the present solutions as well as the return data object of query results are outlined in detail in the documentation.
4 Documentation
The h5rdmtoolbox is versioned via a GitHub repository and can be installed using the Python package installer (pip). At the time of writing, the package version is v1.4.0 and an extensive documentation is automatically created and published online [11]. It provides an overview of features that are not included in this paper or are only briefly discussed.
The documentation website is generated based on Jupyter Notebooks. On the one hand, this results in a practical documentation, showing code and explanations together. On the other hand, it allows users to reuse the code from the documentation for immediate application by simply copying the code snippets. As Jupyter Notebooks become more popular [12], [36], the option to download the full Notebooks will be another efficient option for most users who are new to the toolbox.
5 Limitations
As outlined before, this package serves as a general toolbox, introducing a management layer around HDF5 files. Therefore, its strength lies in the metadata organization and user-friendly interaction with HDF5 files, rather than high-performance data processing. This means, that during dataset creation and reading, additional processing is needed to validate the metadata usage. The process of actually writing and reading the file is dependent on the underlying package, which is h5py. For large datasets, however, the overhead is negligible and the write or read process is dominating. No significant time differences between h5py and h5rdmtoolbox are observed. The same accounts for the performance of working with dataset values. They are provided as xarray objects. Again, generating them based on the numpy array and other information from the HDF5 file requires some time. After this, the performance is dependent on the xarray package.
The chosen design principles introduce two inherent limitations. Firstly, the implementation of the package in Python inherently limits its compatibility for users of other programming languages such as C++, Java or Matlab for instance. The widespread popularity of Python in the scientific community justifies the choice. While a similar implementation in other languages is theoretically possible, such an extension is beyond the scope of this work. Secondly, the selection of HDF5 as the core scientific file format imposes an inherent limitation. Not all scientific or engineering data may be well-suited for HDF5 files. While HDF5 is versatile, some specialized data types or structures may find more suitable alternatives outside the HDF5 format.
Finally, it is essential to note that the number of interfaces to databases and repositories is currently limited. As of the current writing, the database sub-package includes implementations for MongoDB and a query solution using HDF5 itself. In the repository sub-package, only Zenodo is provided. Nevertheless, the toolbox is designed to permit and explicitly encourages further extensions by the community. This open architecture invites collaborative contributions to expand the range of interfaces and integrations with databases and repositories based on the evolving needs and preferences of users.
6 Conclusion and Outlook
The Python package h5rdmtoolbox has been introduced, which is designed to support small collaborative groups, communities, and doctoral researchers who use or consider using HDF5 files as their central file format. HDF5 is chosen for its self-descriptive capabilities and versatility in various scientific domains. However, the management of metadata and internal organization of datasets and groups, as well as facilitating interoperability with other frameworks, is left to the users. The toolbox aims to enhance the FAIR principles of data by providing general, comprehensive tools for managing HDF5 files throughout their lifecycle. While solutions exist, that address management needs, they tend to be domain-specific and lack a generalized framework applicable to diverse problems. Some solutions may only focus on specific aspects of the data lifecycle, such as databases. In contrast, the presented toolbox adopts a broad approach, providing tools that enable users to create tailored management solutions for HDF5 files based on their specific scientific context. Rather than prescribing a singular solution, the toolbox fills the gap between well-established solutions utilized by large scientific communities and the absence of standards for individual researchers. By offering a Python package equipped with high-level tools and interfaces for HDF5 data management, the toolbox improves the FAIRness of HDF5 files for scientists.
With user-friendliness and low entry barriers in mind, the toolbox uses popular Python packages like xarray and pydantic as dependencies and adopts syntax into newly programmed solutions (e.g. query within HDF5 files is adopted from MongoDB). However, the toolbox is missing graphical user interfaces. This would strongly improve the usability and will lower the entry level, especially for less experienced programmers. Future work should set the focus on the design of Conventions and Layouts, as this constitutes the bases of successful data management.
The toolbox has been tested in and improved through various scientific projects with a focus on fluid mechanics. However, further testing in other domains is required. In addition to the implemented unit tests, practical testing in various applications is necessary to identify further needs, weaknesses and thus elaborate potential for improvements. Application to various problems and scientific disciplines are planned and feedback from researchers will need to incorporate into the toolbox. This will extend capabilities, improve the code and allow it to be adapted to the needs of users. Current concrete use cases investigate fluid problems, such as computational fluid dynamics simulations and particle image velocity measurements. Lessons learned from these areas will be incorporated into future publications, while further examples and guidelines will be continuously added to the online documentation [11].
Software availability
Software can be found here
7 Acknowledgements
The software was developed in-house without any external funding and no conflicts of interest are declared. The authors would like to thank all users, who have been testing the toolbox so far and provided helpful feedback. A special thanks belongs to Lucas Büttner for the helpful testing and feedback at the beginning of the project.
8 Roles and contributions
Matthias Probst: Conceptualization, Writing, Software Development – original draft
Balazs Pritz: Project administration, Formal Analysis, Writing - review & editing
References
[1] Y. Wang, Y. Su, and G. Agrawal, “Supporting a Light-Weight Data Management Layer over HDF5,” in 13th IEEE/ACM International Symposium on Cluster, Cloud, and Grid Computing, IEEE, 2013, pp. 335–342. DOI: http://doi.org/10.1109/CCGrid.2013.9.
[2] J. Georgieva, V. Gancheva, and M. Goranova, “Scientific Data Formats,” in Proceedings of the 9th WSEAS International Conference on Applied Informatics and Communications, ser. AIC’09, Moscow, Russia: World Scientific, Engineering Academy, and Society (WSEAS), 2009, pp. 19–24, ISBN: 9789604741076.
[3] E. National Academies of Sciences and Medicine, Open Science by Design: Realizing a Vision for 21st Century Research. Washington, DC: The National Academies Press, 2018. DOI: http://doi.org/10.17226/25116.
[4] F. De Carlo, D. Gürsoy, F. Marone, et al., “Scientific data exchange: a schema for HDF5-based storage of raw and analyzed data,” Journal of synchrotron radiation, vol. 21, no. 6, pp. 1224–1230, 2014.
[5] C. M. Klingner, M. Denker, S. Grün, et al., “Research data management and data sharing for reproducible research—results of a community survey of the german national research data infrastructure initiative neuroscience,” Eneuro, vol. 10, no. 2, 2023.
[6] European Commission and Directorate-General for Research and Innovation, Cost-benefit analysis for FAIR research data : cost of not having FAIR research data. Publications Office, 2019. DOI: http://doi.org/10.2777/02999.
[7] W. K. Michener, “Meta-information concepts for ecological data management,” Ecological Informatics, vol. 1, no. 1, pp. 3–7, 2006. DOI: http://doi.org/10.1016/j.ecoinf.2005.08.004.
[8] N. Preuss, G. Staudter, M. Weber, R. Anderl, and P. F. Pelz, “Methods and technologies for research-and metadata management in collaborative experimental research,” in Applied Mechanics and Materials, Trans Tech Publ, vol. 885, 2018, pp. 170–183.
[9] M. D. Wilkinson, M. Dumontier, I. J. Aalbersberg, et al., “The fair guiding principles for scientific data management and stewardship,” Scientific Data, vol. 3, no. 1, p. 160018, 2016, ISSN: 2052-4463. DOI: http://doi.org/10.1038/sdata.2016.18.
[10] A. Jacobsen, R. de Miranda Azevedo, N. Juty, et al., “FAIR Principles: Interpretations and Implementation Considerations,” Data Intelligence, vol. 2, no. 1-2, pp. 10–29, Jan. 2020, ISSN: 2641-435X. DOI: http://doi.org/10.1162/dint_r_00024.
[11] Probst, Matthias, Documentation of HDF5 Research Data Management Toolbox (v1.4.0), 2024. [Online]. Available: https://h5rdmtoolbox.readthedocs.io/en/v1.4.0/, (accessed: 17.06.2024).
[12] J. M. Perkel, “Why Jupyter is data scientists’ computational notebook of choice,” Nature, vol. 563, no. 7732, pp. 145–147, 2018.
[13] P. Greenfield, M. Droettboom, and E. Bray, “ASDF: A new data format for astronomy,” Astronomy and computing, vol. 12, pp. 240–251, 2015.
[14] E. Taaheri and D. Wynne, “An HDF-EOS and data formatting primer for the ECS project,” Raytheon Company, Tech. Rep., Mar. 2001.
[15] P. Klosowski, M. Koennecke, J. Tischler, and R. Osborn, “NeXus: A common format for the exchange of neutron and synchroton data,” Physica B: Condensed Matter, vol. 241, pp. 151–153, 1997.
[16] T. Hauser, “Parallel i/o for the cgns system,” in nd AIAA Aerospace Sciences Meeting and Exhibit, 2004, p. 1088. DOI: http://doi.org/10.2514/6.2004-1088.
[17] S. Koranne, Hierarchical Data Format 5 : HDF5. Springer, 2011, pp. 191–200, ISBN: 978-1-4419-7719-9. DOI: http://doi.org/10.1007/978-1-4419-7719-9_10.
[18] S. Poirier, A. Buteau, M. Ounsy, et al., “Common Data Model Access: A Unified Layer to Access Data From Data Analysis Point OF View,” Icalepcs, Grenoble, October, 2011.
[19] J. Gregory, “The CF metadata standard,” CLIVAR Exchanges, vol. 8, no. 4, p. 4, 2003.
[20] J. Moore and S. Kunis, “Zarr: A cloud-optimized storage for interactive access of large arrays,” in Proceedings of the Conference on Research Data Infrastructure, vol. 1, 2023.
[21] A. Ingargiola, T. Laurence, R. Boutelle, S. Weiss, and X. Michalet, “Photon-HDF5: an open file format for timestamp-based single-molecule fluorescence experiments,” Biophysical journal, vol. 110, no. 1, pp. 26–33, 2016.
[22] L. Gosink, J. Shalf, K. Stockinger, K. Wu, and W. Bethel, “HDF5-FastQuery: Accelerating complex queries on HDF datasets using fast bitmap indices,” in 18th International Conference on Scientific and Statistical Database Management (SSDBM’06), IEEE, 2006, pp. 149–158.
[23] The HDF Group, Hierarchical Data Format, version 5. [Online]. Available: https://www.hdfgroup.org/HDF5/, (accessed: 18.12.2023).
[24] A. Collette, Python and HDF5. O’Reilly Media, Inc., 2013, ISBN: 9781449367831.
[25] A. Salazar, B. Wentzel, S. Schimmler, R. Gläser, S. Hanf, and S. A. Schunk, “How research data management plans can help in harmonizing open science and approaches in the digital economy,” Chemistry–A European Journal, vol. 29, no. 9, e202202720, 2023.
[26] B. Schembera and D. Iglezakis, “EngMeta: metadata for computational engineering,” International Journal of Metadata, Semantics and Ontologies, vol. 14, no. 1, pp. 26–38, 2020.
[27] D. Iglezakis, D. Terzijska, S. Arndt, et al., “Modelling scientific processes with the m4i ontology,” in Proceedings of the Conference on Research Data Infrastructure, vol. 1, 2023. DOI: http://doi.org/10.52825/cordi.v1i.271.
[28] S. Hoyer and J. Hamman, “xarray: ND labeled arrays and datasets in Python,” Journal of Open Research Software, vol. 5, no. 1, 2017.
[29] Manola, F., Miller, E., Resource Description Framework (RDF). Primer. W3C Recommendation 10 February 2004, 2004. [Online]. Available: http://www.w3.org/TR/rdf-primer/, (accessed: 17.06.2024).
[30] P. Hitzler, M. Krötzsch, S. Rudolph, and Y. Sure, Semantic Web: Grundlagen. Springer, 2008, vol. 1.
[31] D. Brickley and L. Miller, FOAF vocabulary specification 0.99, 2014. [Online]. Available: http://xmlns.com/foaf/spec/, (accessed: 17.06.2024).
[32] S. Arndt, B. Farnbacher, M. Fuhrmans, et al., “Metadata4Ing: An ontology for describing the generation of research data within a scientific activity.,” 2023. DOI: http://doi.org/10.5281/zenodo.8382665.
[33] M.-A. Sicilia, E. García-Barriocanal, and S. Sánchez-Alonso, “Community curation in open dataset repositories: Insights from zenodo,” Procedia Computer Science, vol. 106, pp. 54–60, 2017. DOI: http://doi.org/10.1016/j.procs.2017.03.009.
[34] M. Thelwall and K. Kousha, “Figshare: A universal repository for academic resource sharing?” Online Information Review, vol. 40, no. 3, pp. 333–346, 2016. DOI: http://doi.org/10.1108/OIR-06-2015-0190.
[35] K. Chodorow and M. Dirolf, MongoDB - The Definitive Guide: Powerful and Scalable Data Storage. O’Reilly, 2010, pp. I–XVII, 1–193, ISBN: 978-1-449-38156-1.
[36] T. Kluyver, B. Ragan-Kelley, F. Pérez, et al., “Jupyter Notebooks-a publishing format for reproducible computational workflows.,” Elpub, vol. 2016, pp. 87–90, 2016.