1 Introduction
Research units, from small research groups at universities to large research and development departments are increasingly confronted with the challenge to manage large amounts of data, data of high complexity [1], [2] and changing data structures[3], [4]. The necessary tasks for research data management include storage, findability and long-term accessibility for new generations of researchers and for new research questions[4, 5, 6].
In spite of the advantages of implementing data management solutions [7], we found a lack of standard methods or even standard software so far for research data management, especially in the context of quickly evolving methods and research targets. We hypothesize that the reason for this deficit is that scientific research poses unique challenges for data management, since it is characterized by constant innovation, short lived research questions, trial-and-error approaches, and the continuous integration of new insights.
We propose agile research data management as a promising approach to meet the special requirements of scientific research and to fully leverage the benefits of increased research digitalization, automated data acquisition methods and storage capabilities.
For this article, we identified the specific challenges for research data management (RDM) and defined eleven requirements which suitable RDM software should have to (a) fulfill the practical needs and (b) be accepted by the potential users. We then matched existing tools against these requirements and found areas where the tools show substantial need for improvement.
Finally we present the LinkAhead[8], [9] toolkit as a viable approach to satisfy all the proposed requirements.
2 Challenges for research data management
2.1 The scientific data lifecycle: the need for proper tooling
Data which accrues in scientific research is more than just experimental readings, field notes or interview recordings. In order to fully represent the research journey and eventually enable reproducible science, the data from every research step may become relevant. We identify the challenges to make this data usable in a way that leads to reproducible, and time-efficient, research.
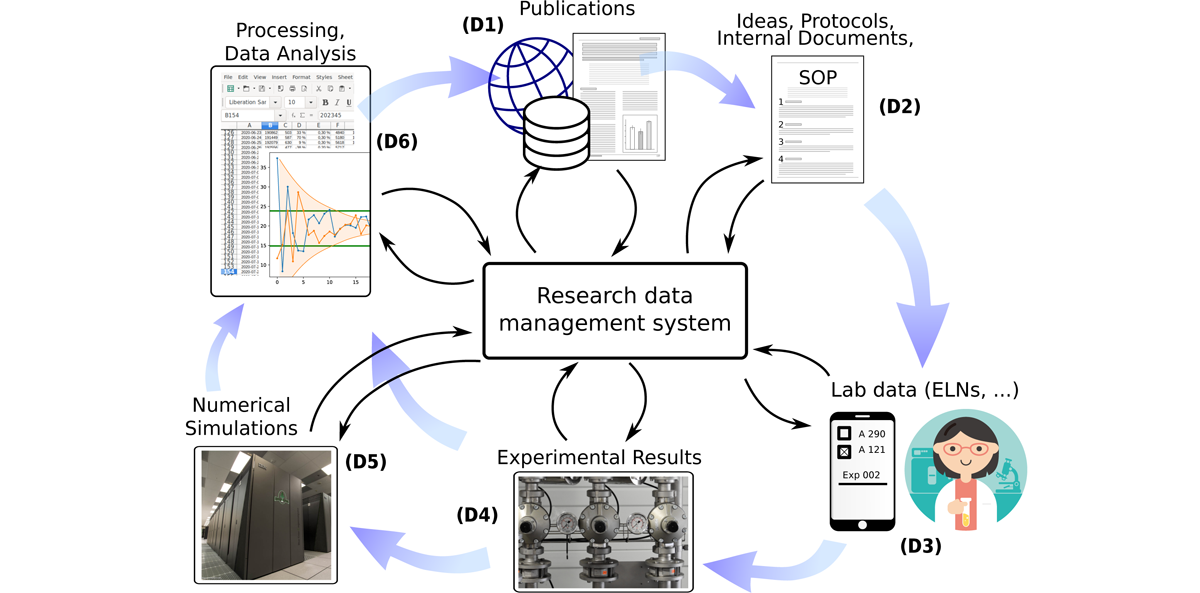
Figure 1 shows a schematic of different research steps during the research lifecycle, during which important data is generated. For full reproducibility, it is not sufficient however to simply store any data that one acquires, but also to represent the semantic connections and make these connections searchable.
Figure 1: Schematic illustration of the scientific data lifecycle. Data can be obtained from every step, and in most cases the relationship between data entities is just as relevant as the raw data. Blue thick arrows denote the direction in which information flows in normal research. Thin black arrows indicate data flow to and from a research data management system. While this example focuses on experimental and laboratory centered disciplines, comparative lifecycles also exist for theoretical sciences and most fields in the humanities.
In more detail, the most relevant sources and targets for data in scientific research are (numbered from (D1) to (D6)):
Prior publications (D1) An important part of good scientific practice (GSP) is to credit the influence of prior work, written by the scientists themselves or third parties. Linking one’s own work to previous publications — articles or published data from repositories — and making these connections public helps to assess reproducibility and may lead to fruitful data-reuse in unforeseen contexts.[10] An RDMS should be able to trace back each data item to previous scientific publications on which it is based.
Ideas and SOPs (D2) The data here consists mostly of text documents which describe thoughts, hypotheses and planned standard operating procedures (SOPs). These documents fill the gap between previous work and the next round of data acquisitions, they often also work as a blueprint for the data acquisition phase.[11] A scientist may consult their RDMS to answer questions like “Which SOP was used when experiment X was carried out to generate data file Y?”.
Lab data (D3) Environmental data, device settings, used SOPs and ingredients and other incidental data typically accrues during the course of experiments and was traditionally stored in paper laboratory notebooks. Currently, a lot of laboratories switch to electronic lab notebooks (ELNs) for the same purpose. While this data is often seen as second-class “metadata”, we hold that since often conclusions can be drawn from it, it deserves the same handling as final instrument readings. [12, 13, 14]
During work in the lab, software must be as unintrusive as possible, with efficient user interfaces.
Experimental results (D4) These are what is often considered the main data. For meaningful analysis, data from experimental results mostly must be enriched with additional data from experimental or device settings or from processed samples, to filter for special conditions, to compare settings or to verify that values are compatible with standard literature.[15]
Numerical simulations (D5) Similarly to experimental results, data obtained from numerical procedures can not be interpreted without knowledge about used software and parameters, possibly hardware conditions and input from laboratories or third-party data sources.[16] Since bit-for-bit reproducibility is possible in theory, all relevant settings should be stored unchanged.
Data analysis (D6) When analyzing data from previous steps, storing not only the used programs, scripts, and their parameters, but also the semantic connections enables later researchers to reconstruct which method was used, which assumptions were made and under which conditions the input data was gathered.[17], [18]
Next publication (D1) Formally the end of the lifecycle, but of course also the beginning of many new ones, a publication contains a number of statements which are supported by data from previous steps. A comprehensive RDMS could quickly answer a question like “In figure X, which methods were used to analyze the data, which devices and software were used to acquire the raw data, and which assumptions were made when planning the experimental setting?”
This list focuses on experimental and laboratory centered disciplines like engineering or natural sciences, but of course in the humanities and theoretical sciences, there are equivalent steps which are equally important to preserve and link to each other.
2.2 Specifics of scientific research data management
There are some needs for data management which are specific to or more pronounced in scientific research, which we will label by (S1) through (S5):
Interoperability Scientists tend to work with their own custom-written software[19, 20, 21], which often requires files with data to be directly accessible to the OS via a file system (S1), remote or locally. Also programmatic access (query, retrieve, update) to data via network APIs (S2) is a necessity for many scientific data uses.
Agility Traditional DMS require users to define a data model and stick to it[22]. All data to be entered has to conform to the data model as it was defined. Research however is defined by having undefined outcomes, the research questions, experimental setup or analysis methods change more often than not over the course of one investigation.[23] We therefore identify (S3) as the special need for flexibility regarding the data model.
Learning curve Scientific research is founded upon the contribution of many participants, with different qualifications, varying research foci and high fluctuations. As a consequence, a steep learning curve for using an RDMS would be detrimental to its adoption (S4).
Early usefulness Systems which only store data, but do not provide short-term advantages, have high acceptance barriers. Especially in academic research, junior scientists with short-term contracts have little incentive to invest time and money in systems which only may pay out on longer timescales.[22] Therefore, RDMS should offer some tangible advantages on the short run (S5).
3 Requirements for a scientific RDMS
Based upon the challenges from the previous section, we propose a set of requirements for an RDMS to be a useful tool for scientific research.
3.1 General requirements
(R1) Semantic linkage In order to retain the semantic context in which data is embedded, it must be possible in the RDMS to link data sets with each other in a meaningful way, i.e., the links must bear some meaning. The default linking possibilities and properties of the data types in the RDMS form the data model.
(R2) Flexible data model Researchers require an RDMS for structured storage of data, where the data model can be changed on the fly, without the need to migrate or discard existing data ((S3)). When the data model is changed, for example due to new machines, protocols or evolving research questions, the existing data must remain valid and usable. A change in ontological semantics now must be compatible with previous semantics then.
(R3) Searchability The RDMS should have easily accessible search options not only for property values of stored entities, but also for links to other entities and properties (and link) thereof. This deep search allows the traversal of the structured knowledge graph and delivers actual utility value.
(R4) Sustainability In order to assure long-term access to stored data, software solutions must have some safeguard against becoming unmaintained. This could be achieved by being either open-source software or “too big to fail”. In the case of open-source software, either the community or other companies could step in, if the original maintainers stopped their support. On the other hand, if a software system is very widely adopted and thus indispensable, it is unlikely to be abandoned or left unsupported.
(R5) Open APIs For interaction with third-party programs, the RDM must have an API with low entrance barriers ((S2)). In research contexts, these third-party programs are often custom-written by scientists without explicit computer science background, so extensive documentation of the API is very desirable.
3.2 Automation
Automation of repetitive data integration reduces error rates and frees users to concentrate on more challenging tasks. It is therefore desirable for an RDMS to have:
(R6) Synchronization The RDMS should make it easy for its administrators to integrate existing data sources (for example databases or file systems with structured folder hierarchies) into the RDMS: The RDMS should be synchronized automatically with data from these sources, which makes these data available in a unified manner via the RDMS interface. Note that the RDMS can not solve the conceptual problem of a single source of truth when synchronizing data from different sources, but it can at least highlight potential conflicts and where they first occurred to administrators.
(R7) ELN integration Research work in the lab is increasingly documented with electronic lab notebooks (ELNs)[24], [25], which allow to conveniently enter device and experimental settings in a semi-structured way. This data is usually critical in the analysis of acquired raw data from instruments, e.g., for searching specific data sets or filtering by parameters. There should be a possibility that the RDMS integrates the ELN data and presents it like data from other sources.
(R8) Workflow representation While following one SOP, the laboratory workflow is often highly standardized, which makes it suitable for representation within the RDMS. The RDMS should support workflows with different states, which can only be switched in an admin-defined pattern. This simplifies the work for users, because they may e.g., only see the interfaces which are relevant for the current sample processing step.
3.3 Specific requirements for scientific work
As introduced in section Specifics of scientific research data management, some requirements arise from scientific research specifically.
(R9) Versioning Mistakes during data acquisition happen, and it must be possible to correct existing data sets. At the same time, this editing must be made transparent and the history of each data set must be kept for future inspection.
(R10) File system integration For interaction with third-party programs, raw data files must be available on standard file systems ((S1)). Ideally the scientists’ workflows should remain unchanged by the RDMS.
(R11) Gentle learning curve, early pay-off To accommodate for the short employment lifecycles in science, RDMS should offer straightforward and simple to learn usage possibilities which give some early sense of achievement ((S4), (S5))[26]. One example could be simplified search options which help users understand that an RDMS will make their work easier when handling with data.
3.4 Relation to FAIR data management
FAIR data management is seen as a general requirement by the scientific community at large. We hold that a research data management system fulfilling (R1) – (R11) can enable research groups to implement a FAIR data management.
Specifically, Findability can be achieved because each data set and collections of data can be assigned persistent identifiers, data and metadata can be intimately connected and data can be found through the search functionality of the RDMS.
Scientific RDMS can enable Accessibility through open and standardized APIs and separation of raw data and metadata. RDMS allow for Interoperability when users can incorporate existing ontologies for data model, descriptions and references between data sets. Reusability is fostered by rich data models including licenses, provenance information and which follow the respective communities’ standards.
4 Current state of the tools landscape
We give a short overview over existing solutions, tools and approaches and over their possibilities. We also classify the extent to which they cover the required features.
4.1 Technologies and approaches
Currently, DMS tools exist for a range of fields and use numerous technological and methodological approaches. Different sources use different definitions for some of the following categories, so we try make our definitions explicit, where necessary.
-
ELNs Here we use the definition of Harvard Medical Scool[27]:
An Electronic Lab Notebook (ELN) is a software tool that in its most basic form replicates an interface much like a page in a paper lab notebook. In an ELN you can enter protocols, observations, notes, and other data using your computer or mobile device.
Electronic laboratory notebooks replace paper-based physical solutions to document the scientific workflow in laboratories, but also partly planning and analysis of obtained data. For the sake of this article, ELNs are distinct from other lab-oriented data management software in that ELNs focus on the user experience while entering data in a laboratory environment and allow to enter data in a semi-structured way, often much like a text editor with the possibiliy to add a number of structured fields. The structure can sometimes be defin by means of user editable templates. Typical examples are eLabFTW[28], Chemotion[29], RSpace[30], eLabJournal[31] and other[24].
Field-specific solutions Many scientific field have specialized data management solutions for their fields which cater to the specific needs, such as chemical structure searches, material property tables, sample management or domain specific data visualization. Often, these solutions excel in their purposes but customization options or interaction possibilities may be limited. Examples are Nomad[32], C6H6.org[33], Chemotion[29], JuliaBase[34] among others.
Data, article and software repositories Most scientific journals and some funding agencies require scientists to publish the data underlying their publications in a publicly accessible data repository. There are data repositories with custom software, and an increasing number of public repository instances using off-the-rack software like Dataverse[35], Invenio[36], DSpace[37] or CKAN[38]. Data repositories cover (D1) in the data lifecycle and offer some search functionality, in all but very few cases they are intended for immutable data at the time of publication. Data models range from very simple (only authors and text description) over completely user defined key-value pairs to domain specific fixed data models for domain repositories.
Similarly, software and articles are stored in specialized repositories, which often have extensive metadata capabilities for the entities stored within them.
Data storage systems Data storage is a necessary prerequisite for scientific research and thus there are many well established systems: mirrored network file systems (e.g., NFS, CIFS) with regular backups to tape archives on the one hand and object stores (e.g., S3) on the other hand, which store binary blobs outside classical file system structures.
SQL databases Plain SQL databases use tables where rows represent records and columns represent the data sets’ attributes or properties. Each table with a fixed set of columns of mostly fixed types represent one type or class of data, defining the properties available for that type.
Because SQL databases are readily available and can be integrated into most programming languages, they are often used as the technical base for both self-written ad-hoc data management solutions and existing commercial data management systems alike[39], [40].
Key-value stores A contrasting approach to SQL databases (therefore categorized as NoSQL databases, popular examples are CouchDB or MongoDB), key-value stores manage data as a collection of key-value pairs. They trade the structure of the SQL paradigm for flexibility, allowing users to store whatever they deem appropriate.
RDF, SPARQL A common concept from academic knowledge representation research, RDF (resource description framework)[41] is a framework and representation standard for subject-predicate-object triples. It has found adoption in the standardization community and some applications. SPARQL is a query language for accessing RDF data and used by knowledge services such as Wikidata.[42], [43]
We would also like to mention that some solutions incorporate one or more if these approaches as components. For example Kadi4Mat[44] and Nomad have ELNs as part of the overall software package.
4.2 Do existing tools meet the requirements?
We discuss to what degree these technologies and tools are able to fulfill the requirements (R1) – (R11) listed above. Here we differentiate between technologies, which may be used when implementing applications on the one hand, and tools on the other hand which are candidates for data management solutions.
4.2.1 Technologies
RDF, SPARQL RDF was designed and is well suited to represent semantic relationships between entities and local RDF collections can be extensively searched with SPARQL by trained experts. There is a number of standardized RDF serializations which can be generated and read by a many programming languages. Data models can be implemented using RDF Schema, which is based upon RDF. Entities can reference entities located on other instances, which brings greater flexibility, but raises issues about data mutability and searchability.
SQL databases Relational databases thrive on relations between tables and thus allow some degree of semantic linking, albeit with very limited flexibility. Searching is possible, but requires a certain degree of expertise, which can be mitigated by external helper tools. There are standardized implementations, open source and proprietary alike, which can be expected to continue for the foreseeable future.
Key-value stores NoSQL databases allow users a comprehensive degree of freedom when storing data, but at the same time often provide no overarching structure to enforce certain data model properties. Semantic linkage thus often is limited to convention instead of internalized structures. Searchability is comparable to traditional SQL databases, and there is a large number of implementations.
Data storage systems As a basic technology to store raw files or objects, data storage systems do not have the ability to link data or provide a data model. Searching data for associated metadata or file content is possible for some storage systems. Higher-level functionality is not available within the data storage systems themselves.
These base technologies have in common that they mostly do not provide functionality such as high-level network APIs, graphical user interfaces, integration with other components or versioning. Also they target technical audiences and thus feature steep learning curves for data manipulation and searching alike.
Table 1: Data technologies, tools and if they meet the requirements.
| Technology | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | R10 | R11 |
| RDF + SPARQL | ● | ● | ● | ● | ● | ○ | ∅ | ∅ | ○ | ○ | ○ |
| SQL | ● | ○ | ● | ● | ● | ○ | ∅ | ∅ | ○ | ○ | ○ |
| Key-value stores | ◐ | ◐ | ◐ | ● | ● | ○ | ∅ | ∅ | ○ | ○ | ○ |
| Data storage | ○ | ○ | ◐ | ● | ● | ∅ | ∅ | ∅ | ◐ | ● | ◐ |
| Tools | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | R10 | R11 |
| ELNs | ◐ | ◐ | ◐ | ● | ● | ◐ | ∅ | ● | ● | ◐ | ● |
| FSSa | ◐ | ◐ | ◐ | ◐ | ◐ | ☉ | ☉ | ● | ◐ | ◐ | ● |
| Repositories | ○ | ○ | ◐ | ● | ◐ | ☉ | ○ | ○ | ● | ○ | ● |
| a. field-specific solutions | |||||||||||
| R1 Semantic linkage | R7 ELN integration | ||||||||||
| R2 Flexible data model | R8 Workflow representation | ||||||||||
| R3 Searchability | R9 Versioning | ||||||||||
| R4 Sustainability | R10 File system integration | ||||||||||
| R5 Open APIs | R11 Gentle learning curve, early pay-off | ||||||||||
| R6 Synchronization | |||||||||||
Symbols used: ●:yes, ○: no, ◐: partly, ☉: may be possible to implement, ∅: not applicable. Note that a “◐” may signify that not all particular examples of the category fulfill the requirement, but it may also mean that (nearly) all examples fulfill parts of the requirement.
4.2.2 Tools
-
ELNs ELNs target at a non-technical audience and thus generally aim to have low entrance barriers, with tutorials and graphical help functions. Most generic ELNs allow basic linking between stored records and searches thereof, and users are guided in their work of entering data by means of templates. These templates often do not have a semantic meaning however, but serve only as a means of suggesting data fields. Data is organized around lab sessions, the main datatype are notes from the laboratory. ELNs only started to become the de-facto standard in laboratories over the last decade, so the market is far from settled. There are open-source and proprietary software solutions, by large players and by solo enterprises. Nearly all ELNs developed over the last five years now offer APIs for third-party access, and many allow users to organize their workflows, such as different processing steps for a sample.
Synchronization with other data sources or integration with file systems is not a core element of ELNs and as such rarely seen. Similarly, synchronization with other data sources exists only on a case-to-case base. Versioning of stored entities is possible to some extent for most ELNs.
Field-specific solutions Semantic linking may be possible to a certain amount as permitted by the data model, which typically is limited to the use cases foreseen by the developers. Similarly, searching the data often is limited to key-value filters on the specialized data types. Some solutions (e.g., NOMAD) implement their own ELNs, but integration with third-party ELNs and synchronization with other data sources does not exist generally: it could be implemented via APIs, in those cases where they exist. Support for workflows is generally quite good, and the learning curves are adapted to the audience. Versioning of data and integration of existing file systems may be present in some systems. Long-term availability of software support may be an issue when these solutions are only developed by a small set of people or even individuals, often in time-limited funding situations. In these cases, open-source software can be an insurance for the future, especially if there is sufficient development documentation.
Repositories Data repositories only cover a small subset of data management use cases and as such generally do not implement many of the requirements. They may allow semantic linkage between entities, but do have encompassing data models at all. Searching is limited to key-value filters and full-text, sometimes referenced data sets can also be used as filters, but there may be APIs which allow external tools to improve on this shortcoming. Repositories generally have institutional funding so that long-term availability can be seen as guaranteed. Synchronization with other data sources, local file system or ELN integration or workflow representation does not make sense, since repositories are meant for manual data archive upload at the end of the scientific life cycle. Upload of data to archives is very straightforward in most cases, and editing of uploaded data does not invalidate the original version, but only marks it as out of date.
4.2.3 Summary of existing tools
The requirements coverage of the examined technology and tool classes are shown in table 4.2.1. We see that while existing tools cover a wide range of the required features, there are significant shortcomings in two areas: flexible data models, semantic linkage and searchability on the one hand, and integration with ELNs, other devices and file systems on the other hand. Due to the large number of available products, for each requirement, there are ELN and field-specific solutions which may fulfill it at least partly, although such a product in general does not cover all requirements.
We stated earlier that these topical fields are especially relevant in scientific research. As an effect DMS have been widely successful in many areas such as finance, administration, and high-tech industries[45], [46], but remain scarce in both academic and private sector research[46], [47]. In summary, we find the need for a tool which fills the requirements for semantic, flexible data management and has sufficient synchronization and ELN integration capabilities.
5 LinkAhead
We hold that LinkAhead[9], an agile data management framework, fulfills the proposed requirements from section Requirements for a scientific RDMS. LinkAhead was initially developed under the name “CaosDB” by one of our colleagues, Timm Fitschen, during his time at the Max Planck Institute for Dynamics and Self-Organization, and others.[48] In 2018, LinkAhead was released, still as “CaosDB” under the AGPLv3 license on gitlab.com.[8] Since 2020, LinkAhead has found increased adoption in multiple research facilities.
In this section, we first describe LinkAhead in detail, then we assess to which extent LinkAhead fulfills the proposed requirements and finally we give an overview over limitations and possible enhancements in the future.
5.1 Detailed description
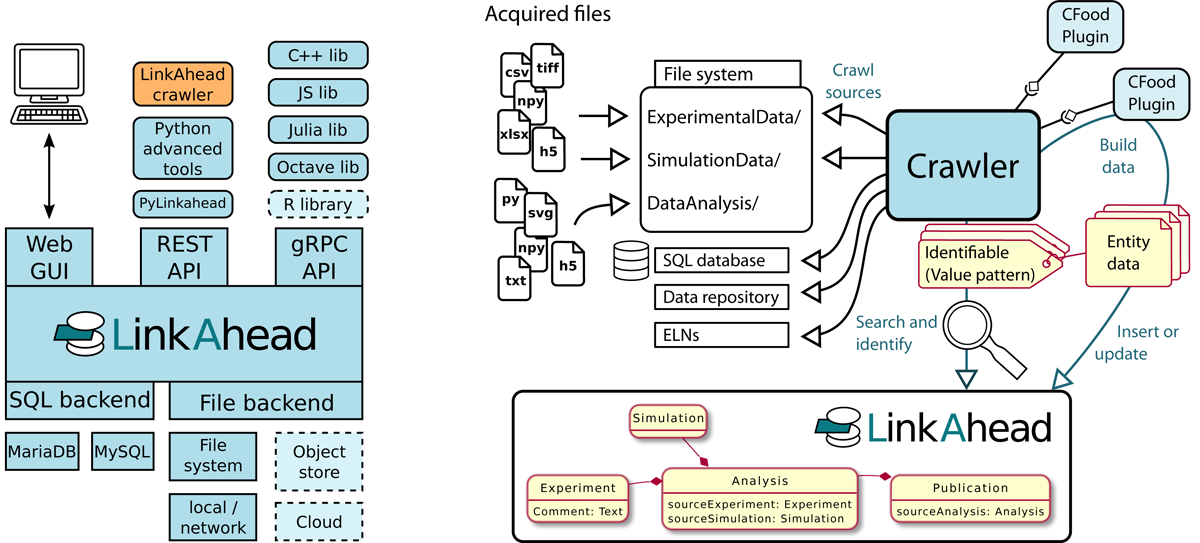
LinkAhead was developed out of the need for a data management solution that can cope with large data amount from automated sources and from existing file systems alike and that allows researchers to quickly adapt the way how data sets are connected or described. These needs reflect on the design choices which were taken over the course of development. LinkAhead is a general research data management system: specialized solutions such as ELNs, sample management systems, document management systems or other can be developed on top of it, according to specific needs.
5.1.1 Data Model
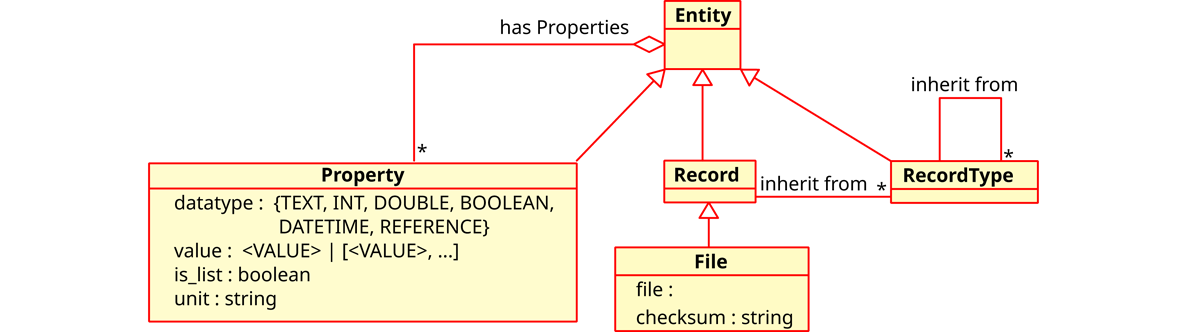
LinkAhead’s meta data model is shown schematically in Figure 2. The base type for everything is Entity, with the inheriting types Property (attributes of Entities, may be list values and references to other Entities), RecordType (templates for actual data sets) and Record. Actual data is typically stored in Records, which inherit from one or more RecordTypes and thus have all the Properties defined therein. The RecordTypes may form a complex inheritance hierarchy themselves. File entities are similar to Records, but additionally are connected to files which may reside on conventional file systems or potentially in abstracted cloud storage systems. This approach to use files at their current locations instead of duplicating file content not only increases LinkAhead’s scalability, but also lower the entrance barrier, since scientists can access the managed file in their traditional ways.
Details of this metadata model in LinkAhead are elaborated on in [9], but it should be clear now already that LinkAhead provides the Semantic linkage feature.
In LinkAhead, the data model of the stored data refers to the RecordTypes and their Properties, which together describe the pattern to which newly created data sets should conform. The data model in LinkAhead can be modified at any time, but the changes only take effect for data to be inserted after this modification. Existing data is not affected and remains unchanged. This property fulfills the proposed Flexible data model feature.
Properties of RecordTypes are allocated a graded importance, which denotes if this Property is either obligatory, recommended or merely suggested for Records which inherit from this RecordType, when a user creates a new Records. This system of importances and the fact that legacy data is not necessarily consistent with a modified data model was a deliberate design decision. The rationale was that when the data model changes, the meaning at the time of data creation should have priority over consistency with later data models.
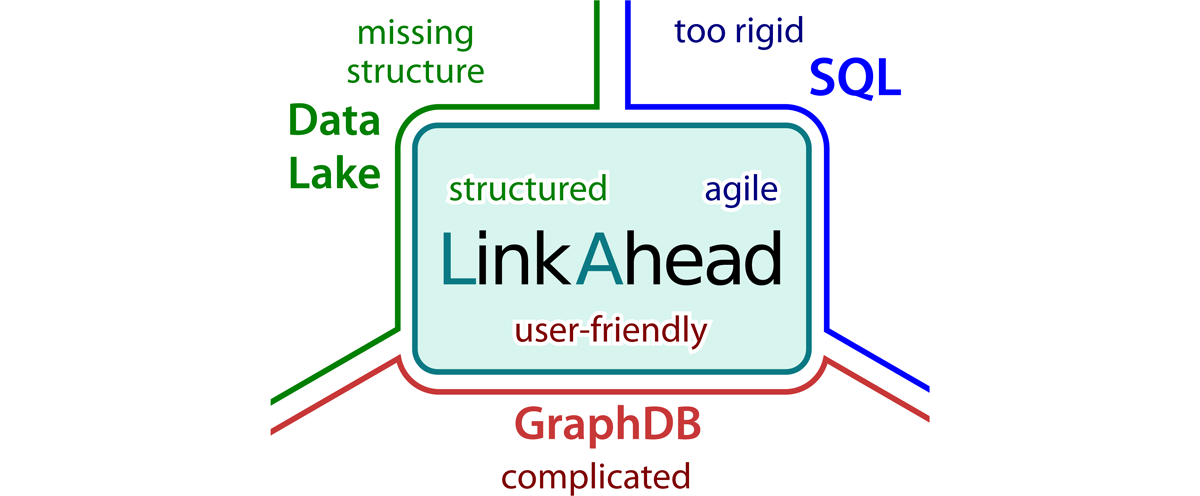
This possibility to completely change the data model, while not giving up on a general structure, places LinkAhead between traditional SQL based relational databases and NoSQL approaches (c.f. Figure 3). While we described above why rigid SQL databases are not suited for use in dynamic research environments, giving no structure (the NoSQL paradigm) tends to lead to incoherent data which is hard to search. A common implementation of NoSQL approaches in the context of data management are data lakes, where raw data can be stored and annotated with metadata. The missing structure in Data Lakes however has lead to the tongue-in-cheek colloquialism “Data Swamp”. A third approach, using graph databases to represent semantic information, has not found its way into general adoption to our knowledge, presumably because the query languages tend to become very unwieldy, compare the appendix Appendix: Query language comparison for an example.
5.1.2 Architecture and Libraries
LinkAhead uses a client/server based architecture, as depicted in Figure 4a. LinkAhead has is a REST API for simple access by traditional clients and a web interface for browsers, as well as a gRPC API which allows for more complex operations, such as atomic content manipulations. The existing client libraries1 and the open APIs provide the proposed Interoperability requirement.
One particularly useful client library component is the LinkAhead Crawler framework. This extensible framework simplifies the work to synchronize external data sources with LinkAhead through a plugin system. The crawler workflow can be characterized as follows:
The crawler checks its data sources for new or changed data stores, such as file systems or the content of other databases. This may happen periodically or be triggered manually by users.
Each new data source is fed to a so-called CFood plugin for consumption. There is a choice of existing plugins, or administrators can write their own. The CFood plugin’s job is to build LinkAhead entities from the consumed data and to specify Identifiables, which work as search patterns. Administrators can mostly define simple CFood plugins by YAML configuration files[49] which is a more user-friendly approach than for example the mappings defined by the W3C’s R2RML standard.[50]
The crawler checks for each Identifiable if a corresponding entity exists already in LinkAhead. If there is no corresponding entity, the entity as returned by the CFood plugin is inserted into LinkAhead. If there is already an existing entity, the Crawler will attempt to merge the existing with the new entity and notify the data curators in case of merge conflicts.
This tool set provides the Synchronization requirement, and if ELNs are used as external data source, the ELN integration. Practical use of LinkAhead crawler framework has previously been demonstrated in [51] and ELN integration was implemented as a working proof-of-concept in [52].
5.1.3 Miscellaneous features
-
Deep search LinkAhead offers a simple semantic query language, which borrows some semantics from SQL, but has a focus on usability for non-technical users. The LinkAhead query language makes deep search easy with expressions like the following:
FIND Analysis WITH quality_factor > 0.5 AND WITH Sample WITH weight < 80g This convenient nesting of query expressions circumvents the JOIN operations from traditional SQL languages. A full documentation of LinkAhead’s query language is available online[53] and in LinkAhead’s sources.
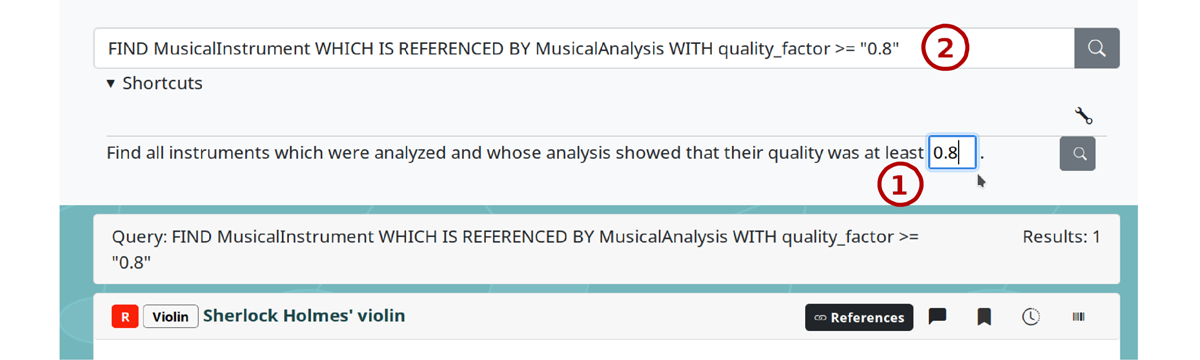
Search templates LinkAhead’s web interface provides customizable search templates which allow more advanced users to create their own query templates, which can then be shared with novice users for simplified searches. In query templates, users can insert custom strings into pre-defined locations of a search query, see Figure 5.
Versioning When entities are modified in LinkAhead, time and user of the change are recorded and LinkAhead puts the previous version onto a history stack and amends the current version with link to the previous version. Over time, each entity may thus grow to a tree of linked versions, which can be retrieved via the web UI or programmatically through the APIs. This feature of LinkAhead enables scientific research data management users to adhere to the principles of good scientific practice.
State management In LinkAhead, users may declare a state machine of states and allowed transitions. Users may then affix states to entities, and these states can then only be changed according to the rules of the state machine. In this way, users can implement a workflow representation which ensure that for example laboratory samples run through a specified list of preparation steps in order.
Figure 5: A query template in LinkAhead’s web UI. The user can enter a custom value into an input field ① and the template is then executed as a plain LinkAhead query ②. Screenshot from https://demo.indiscale.com.
5.1.4 Availability and documentation
LinkAhead is available on the public Git repository gitlab.com at https://gitlab.com/linkahead, a detailed list of LinkAhead’s sub projects is given in the annex. LinkAhead’s source code is licensed under the AGPLv3 (Affero GNU Public License, version 3). Community contribution workflows, a code of conduct and general development guidelines are outlined in https://gitlab.com/linkahead/linkahead-meta and in the sub project specific code repositories. The community chat[54] is currently populated with 33 members, the contributors file lists 19 active contributors[48].
For the interested public, there is a live demo server at https://demo.indiscale.com, hosted by IndiScale GmbH. IndiScale GmbH also provides commercial support, development and customization services for LinkAhead. There are also Debian/Ubuntu packages to run precompiled LinkAhead for download at https://indiscale.com/download.
LinkAhead’s sub projects each have their own documentation in their source directories. The documentation is also available online at https://docs.indiscale.com.
5.2 Requirements matching
In the following list, we evaluate if and how LinkAhead matches the requirements proposed in section Requirements for a scientific RDMS:
(R1) Semantic linkage Links between Entities in LinkAhead are implemented as reference typed Properties, these Properties can be restricted to Entities with certain parents, adding an additional ontological level. All Properties can have a description and higher-order properties and thus can fulfill the requirements for typical predicates in subject-predicate-object relationships in predicate logic oriented triple stored such as RDF.
(R2) Flexible data model In LinkAhead, the data model, i.e., the set of RecordTypes can be modified at any time. Existing Records are not affected by these modifications and keep their properties and inheritance information.
(R3) Searchability LinkAhead’s query language allows to deeply search the available data for simple key-value relations and also for nested relations on the knowledge graph and the related entities’ properties.
(R4) Sustainability LinkAhead is fully open-source and freely available on gitlab.com, with options for commercial support.
(R5) Open APIs The REST and GRPC APIs included in LinkAhead enable interaction with scientists’ custom-written programs. Additionally the existence of client libraries simplifies the usage by programmers without formal software development training.
(R6) Synchronization LinkAhead’s crawler framework simplifies the synchronization between existing data sources and the RDMS and allows to make a diversity of data accessible at a single resource.
(R7) ELN integration The LinkAhead crawler may use ELNs as a data source, thus integrating the content acquired by ELNs into the RDMS. This makes ELN data searchable and usable equivalently to data from their sources.
(R8) Workflow representation The state machine in LinkAhead can be used to represent standardized workflows. For example laboratory samples or interview partners or publications may have a state whose possible transitions and conditions can be specified.
(R9) Versioning Entities in LinkAhead are versioned and previous content may be displayed and recovered. The content history of entities is stored: which user changed what value at which time.
(R10) File system integration LinkAhead does not make copies of data files but only references the file locations. The file path or resource identifier is returned upon queries, so that users can use the location in their accustomed software.
(R11) Gentle learning curve, early pay-off Search queries in LinkAhead can be made more accessible to users by templates where only specific values need to be filled in. The agile data model allows scientists to start with a structured data management without the need to develop a seemingly overwhelming master plan for their data. Instead they can start small in an area where they expect the most immediate benefits such as improved findability of linked data, and grow the data management at a later time.
We find that LinkAhead fulfills the requirements (R1)–(R5), (R9)–(R11) “out of the box” and that (R6)–(R8) (synchronization, ELN integration and workflows) can be readily implemented using on-board means. LinkAhead therefore qualifies as a promising candidate for a scientific RDMS.
5.3 Critical evaluation and outlook
A common misunderstanding about LinkAhead is what it provides out of the box and what it can be used for. LinkAhead is not a tool to describe data objects following a specific ontology, but ontologies can be implemented with LinkAhead in a straightforward manner, and it makes it easy to manage data according to that ontology. It is not an ELN either: ELNs focus on unintrusive interfaces for manual data acquisition, but mostly leaving handling of data from other sources, or semantic data searches, to other tools. LinkAhead can be seen as a perfect complement to ELNs, its primary goal is to make searching and linking of data beneficial for its users and to allow for automation of all tasks. One data source for this automation may be ELNs, but of course also other scientific data acquisition appliances such as laboratory hardware, high-performance clusters or data repositories.
Similarly, LinkAhead does not enforce data to be FAIR. However researchers can use LinkAhead to implement a FAIR data management and to assure that they handle their data in a FAIR manner. Data transferred over the REST and GRPC interfaces use standardized formats such as XML for data serialization, which can be understood by most programming interfaces. Additionally, the internal infrastructure of LinkAhead is being reworked to use UUIDs or other unique identifiers as primary keys for all Entities.
As outlined in the previous section, LinkAhead fulfills most of the requirements and makes others feasible for administrators and users. This also implies that there is room for improvement, for example by providing integrated connectors to ELNs or other data sources or templates for workflow representations.
Along similar lines, LinkAhead is still lacking tools to seamlessly interchange data and data models with RDF based systems. In order to accelerate the general interoperability between data management tools, LinkAhead has become part of the ELN consortium[55], an association of interested parties with the aim to develop a common interchange format, based upon the RO-Crate[56], [57] specification. While it is possible now already by external tools, full integration of existing vocabularies represented in RDF serializations will further simplify FAIR data handling with LinkAhead.
When synchronizing data with LinkAhead, special attention has to be given to the relationship between data from external sources (e.g., crawled files, ELNs) and records in the RDMS. Different sources can (usually by some error) have conflicting data, or entries in the RDMS can be changed manually by users after their insertion. In our experience, this problem can not be solved in a general and purely technical way. Instead, best practices have to be implemented as to where possible errors should be corrected and whether some sources have precedence above each other. An RDMS like LinkAhead, together with the crawler framework, can help administrators identify inconsistencies in the case of two or more data sources. Through versioning, it is visible who and when maybe changed data manually. How to optimize the help in recognizing potential conflicts, and in the end curate data both in the RDMS and in the external sources, is subject of the authors’ ongoing research.
Since LinkAhead does not receive institutional funding, the direction of its future development depends on the actions of the community. Therefore the immediate advancements will be shaped by the needs of the current users of LinkAhead and of the company which currently provides commercial support for it. A current list of feature requests can be generated online.[58] The authors know of about a dozen institutions where LinkAhead is currently in use. Together with the growing user base we expect the software to persist for a significant amount of time.
LinkAhead may fall short in terms of performance against traditional SQL databases for very large amounts of data. To address this issue there is currently development underway to add a virtualization layer which may use existing tabular data sources and present them in a configurable way as native LinkAhead Entities.[59]
We are aware that the perceived “usability” is subject to personal preferences unless evaluated in a controlled study. We see the potential for a separate survey in the future which systematically evaluates user experiences, workflows and the time and effort spent or gained by users of different software approaches to a previously defined set of data management challenges.
6 Conclusion
We found that scientific research has specific needs to data management: Interoperability, agility, adequate learning curves and early practical use. Altogether we identified a set of eleven requirements which we applied to multiple classes of technologies and tools and to LinkAhead, an agile RDMS. Especially in the requirements cluster “Semantic linkage, flexible data model, semantic search”, previously existing tools show significant weaknesses, whereas LinkAhead offers a promising outlook.
We hope that the open source license of LinkAhead will inspire more scientists to contribute to LinkAhead and improve it in the areas of interoperability with existing standards.
7 Appendix: Software
7.1 LinkAhead
The LinkAhead suite with the main libraries is published at Zenodo:
https://zenodo.org/record/7752417 (DOI: 10.5281/zenodo.7752417)
7.2 List of LinkAhead libraries
The following libraries for programming client applications are publicly available:
-
Python https://gitlab.com/linkahead/linkahead-pylib The Python client library can be used for third-party applications and is the foundation for several other libraries:
Advanced Python tools https://gitlab.com/linkahead/linkahead-advanced-user-tools Additional high-level tools building upon the Python library, including a legacy implementation of the LinkAhead crawler. These tools also include converters from JSON Schema to LinkAhead’s data model.
Crawler https://gitlab.com/linkahead/linkahead-crawler A new implementation of the LinkAhead crawler, also using the Python library. Allows to validate data against a JSON Schema.
JavaScript https://gitlab.com/linkahead/linkahead-webui The JavaScript library is part of the web user interface component.
Protobuf API https://gitlab.com/linkahead/linkahead-proto The gRPC API is defined via these Protobuf files.
C++ https://gitlab.com/linkahead/linkahead-cpplib The C++ library uses the gRPC API of LinkAhead.
Octave https://gitlab.com/linkahead/linkahead-octavelib The Octave/Matlab library is based upon the C++ library.
Julia https://gitlab.com/linkahead/linkahead-julialib The Julia library also is based upon the C++ library.
8 Appendix: Query language comparison
As an example for nested queries in different query languages, we consider the search for female UK-based writers in a certain time period, whose given or family name starts with the letter “M”. We used the RDF query language SPARQL with Wikidata (https://www.wikidata.org) identifiers and LinkAhead’s query language with fictional but realistic identifier names.
The SPARQL query is as follows:
1 SELECT DISTINCT ?item ?itemLabel ?givenName ?familyName WHERE {
2 ?item wdt:P31 wd:Q5; # Any instance of a human.
3 wdt:P27 wd:Q145; # citizenship in the United Kingdom
4 wdt:P21 wd:Q6581072; # female
5 wdt:P106 wd:Q36180; # writer
6 wdt:P569 ?birthday;
7 wdt:P570 ?diedon;
8 wdt:P734 [rdfs:label ?familyName];
9 wdt:P735 [rdfs:label ?givenName].
10 FILTER(?birthday > "1870-01-01"^^xsd:dateTime
11 && ?diedon < "1950-01-01"^^xsd:dateTime)
12 FILTER(regex (?givenName, "M.*") || regex (?familyName, "M.*"))
13 SERVICE wikibase:label { bd:serviceParam wikibase:language "en" }
14 }
In contrast, the LinkAhead query looks like this:
1 SELECT given_name, family_name FROM Writer
2 WITH gender=f AND citizenship=UK AND birthday > 1870 AND death < 1950
3 AND (given_name LIKE "M*" OR family_name LIKE "M*")
We understand that SPARQL and LinkAhead’s query language have non-overlapping sets of features. For example, LinkAhead does not know about aliases for names, such as in multilingual environments. On the other hand, SPARQL has no native understanding of SI units and their conversion and it focuses on experts instead of casual users.
Data Availability
No data was used for this article.
Software Availability
Software can be found here: https://gitlab.com/linkahead and at DOI: 10.5281/zenodo.7752417
Notes
- A list of the available libraries with the respective source code repositories are given in the Appendix section List of LinkAhead libraries. [^]
9 Acknowledgements
We acknowledge the previous work on the LinkAhead software by its main authors and independent contributors[48], especially Timm Fitschen  .
.
10 Conflicts of interest
The authors work for IndiScale GmbH, which provides commercial support and other services for LinkAhead. DH and FS contributed to the development of LinkAhead.
11 Roles and contributions
Daniel Hornung: Conceptualization, Visualization, Writing – original draft
Florian Spreckelsen: Conceptualization, Writing – review & editing
Thomas Weiß: Conceptualization, Visualization
References
[1] C. R. Bauer, N. Umbach, B. Baum, et al., “Architecture of a Biomedical Informatics Research Data Management Pipeline,” Exploring Complexity in Health: An Interdisciplinary Systems Approach, pp. 262–266, 2016. doi: http://doi.org/10.3233/978-1-61499-678-1-262. [Online]. Available: https://ebooks.iospress.nl/doi/10.3233/978-1-61499-678-1-262.
[2] C. L. Borgman, “The conundrum of sharing research data,” Journal of the American Society for Information Science and Technology, vol. 63, no. 6, pp. 1059–1078, 2012, issn: 1532-2890. doi: http://doi.org/10.1002/asi.22634. [Online]. Available: https://onlinelibrary.wiley.com/doi/abs/10.1002/asi.22634.
[3] A. M. Khattak, K. Latif, and S. Lee, “Change management in evolving web ontologies,” Knowledge-Based Systems, vol. 37, pp. 1–18, Jan. 1, 2013, issn: 0950-7051. doi: http://doi.org/10.1016/j.knosys.2012.05.005. [Online]. Available: https://www.sciencedirect.com/science/article/pii/S0950705112001323 (visited on 01/18/2023).
[4] T. Schneider and M. Šimkus, “Ontologies and Data Management: A Brief Survey,” KI -Künstliche Intelligenz, vol. 34, Aug. 13, 2020. doi: http://doi.org/10.1007/s13218-020-00686-3.
[5] M. D. Wilkinson, M. Dumontier, IJ. J. Aalbersberg, et al., “The FAIR Guiding Principles for scientific data management and stewardship,” Scientific Data, vol. 3, no. 1, p. 160 018, 1 Mar. 15, 2016, issn: 2052-4463. doi: http://doi.org/10.1038/sdata.2016.18. [Online]. Available: https://www.nature.com/articles/sdata201618 (visited on 11/22/2021).
[6] R. Higman, D. Bangert, and S. Jones, “Three camps, one destination: The intersections of research data management, FAIR and Open,” Insights, vol. 32, no. 1, p. 18, 1 May 22, 2019, issn: 2048-7754. doi: http://doi.org/10.1629/uksg.468. [Online]. Available: http://insights.uksg.org/articles/10.1629/uksg.468/ (visited on 01/18/2023).
[7] G. W. Hruby, J. McKiernan, S. Bakken, and C. Weng, “A centralized research data repository enhances retrospective outcomes research capacity: A case report,” Journal of the American Medical Informatics Association, vol. 20, no. 3, pp. 563–567, May 1, 2013, issn: 1067-5027. doi: http://doi.org/10.1136/amiajnl-2012-001302. [Online]. Available: https://doi.org/10.1136/amiajnl-2012-001302.
[8] “LinkAhead.” Website: https://getlinkahead.com, Source code: https://gitlab.com/linkahead, GitLab. (Oct. 5, 2023).
[9] T. Fitschen, A. Schlemmer, D. Hornung, H. tom Wörden, U. Parlitz, and S. Luther, “CaosDB — Research Data Management for Complex, Changing, and Automated Research Workflows,” Data, vol. 4, no. 2, p. 83, 2 Jun. 2019, issn: 2306-5729. doi: http://doi.org/10.3390/data4020083. [Online]. Available: https://www.mdpi.com/2306-5729/4/2/83 (visited on 01/18/2023).
[10] F. Radicchi, S. Fortunato, and A. Vespignani, “Citation Networks,” in Models of Science Dynamics: Encounters Between Complexity Theory and Information Sciences, ser. Understanding Complex Systems, A. Scharnhorst, K. Börner, and P. van den Besselaar, Eds., Berlin, Heidelberg: Springer, 2012, pp. 233–257, isbn: 978-3-642-23068-4. doi: http://doi.org/10.1007/978-3-642-23068-4_7. [Online]. Available: https://doi.org/10.1007/978-3-642-23068-4_7.
[11] K. Manghani, “Quality assurance: Importance of systems and standard operating procedures,” Perspectives in Clinical Research, vol. 2, no. 1, pp. 34–37, 2011, issn: 2229-3485. doi: http://doi.org/10.4103/2229-3485.76288. pmid: 21584180. [Online]. Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3088954/ (visited on 07/11/2023).
[12] M. A. Abujayyab, M. Hamouda, and A. Aly Hassan, “Biological treatment of produced water: A comprehensive review and metadata analysis,” Journal of Petroleum Science and Engineering, vol. 209, p. 109 914, Feb. 1, 2022, issn: 0920-4105. doi: http://doi.org/10.1016/j.petrol.2021.109914. [Online]. Available: https://www.sciencedirect.com/science/article/pii/S0920410521015308 (visited on 07/11/2023).
[13] A. Nicholson, D. McIsaac, C. MacDonald, et al., “An analysis of metadata reporting in freshwater environmental DNA research calls for the development of best practice guidelines,” Environmental DNA, vol. 2, no. 3, pp. 343–349, 2020, issn: 2637-4943. doi: http://doi.org/10.1002/edn3.81. [Online]. Available: https://onlinelibrary.wiley.com/doi/abs/10.1002/edn3.81.
[14] J. Griss, Y. Perez-Riverol, H. Hermjakob, and J. A. Vizcaíno, “Identifying novel biomarkers through data mining—A realistic scenario?” PROTEOMICS – Clinical Applications, vol. 9, no. 3–4, pp. 437–443, 2015, issn: 1862-8354. doi: http://doi.org/10.1002/prca.201400107. [Online]. Available: https://onlinelibrary.wiley.com/doi/abs/10.1002/prca.201400107.
[15] G. Silvello, G. Bordea, N. Ferro, P. Buitelaar, and T. Bogers, “Semantic representation and enrichment of information retrieval experimental data,” International Journal on Digital Libraries, vol. 18, no. 2, pp. 145–172, Jun. 1, 2017, issn: 1432-1300. doi: http://doi.org/10.1007/s00799-016-0172-8. [Online]. Available: https://doi.org/10.1007/s00799-016-0172-8.
[16] B. G. Fitzpatrick, “Issues in Reproducible Simulation Research,” Bulletin of Mathematical Biology, vol. 81, no. 1, pp. 1–6, Jan. 1, 2019, issn: 1522-9602. doi: http://doi.org/10.1007/s11538-018-0496-1. [Online]. Available: https://doi.org/10.1007/s11538-018-0496-1.
[17] R. A. Poldrack, K. J. Gorgolewski, and G. Varoquaux, “Computational and Informatic Advances for Reproducible Data Analysis in Neuroimaging,” Annual Review of Biomedical Data Science, vol. 2, no. 1, pp. 119–138, 2019. doi: http://doi.org/10.1146/annurev-biodatasci-072018-021237. [Online]. Available: https://doi.org/10.1146/annurev-biodatasci-072018-021237.
[18] F. Strozzi, R. Janssen, R. Wurmus, et al., “Scalable Workflows and Reproducible Data Analysis for Genomics,” in Evolutionary Genomics: Statistical and Computational Methods, ser. Methods in Molecular Biology, M. Anisimova, Ed., New York, NY: Springer, 2019, pp. 723–745, isbn: 978-1-4939-9074-0. doi: http://doi.org/10.1007/978-1-4939-9074-0_24. [Online]. Available: https://doi.org/10.1007/978-1-4939-9074-0_24.
[19] J. E. Hannay, C. MacLeod, J. Singer, H. P. Langtangen, D. Pfahl, and G. Wilson, “How do scientists develop and use scientific software?” In 2009 ICSE Workshop on Software Engineering for Computational Science and Engineering, May 2009, pp. 1–8. doi: http://doi.org/10.1109/SECSE.2009.5069155.
[20] J. Segal, “When Software Engineers Met Research Scientists: A Case Study,” Empirical Software Engineering, vol. 10, no. 4, pp. 517–536, Oct. 1, 2005, issn: 1573-7616. doi: http://doi.org/10.1007/s10664-005-3865-y. [Online]. Available: https://doi.org/10.1007/s10664-005-3865-y.
[21] G. Wilson, “Software Carpentry: Getting Scientists to Write Better Code by Making Them More Productive,” Computing in Science & Engineering, vol. 8, no. 6, pp. 66–69, Nov. 2006, issn: 1558-366X. doi: http://doi.org/10.1109/MCSE.2006.122.
[22] N. R. Anderson, E. S. Lee, J. S. Brockenbrough, et al., “Issues in Biomedical Research Data Management and Analysis: Needs and Barriers,” Journal of the American Medical Informatics Association, vol. 14, no. 4, pp. 478–488, Jul. 1, 2007, issn: 1067-5027. doi: http://doi.org/10.1197/jamia.M2114. [Online]. Available: https://doi.org/10.1197/jamia.M2114.
[23] M. Bron, J. Van Gorp, and M. de Rijke, “Media studies research in the data-driven age: How research questions evolve,” Journal of the Association for Information Science and Technology, vol. 67, no. 7, pp. 1535–1554, 2016, issn: 2330-1643. doi: http://doi.org/10.1002/asi.23458. [Online]. Available: https://onlinelibrary.wiley.com/doi/abs/10.1002/asi.23458.
[24] S. Kanza, C. Willoughby, N. Gibbins, et al., “Electronic lab notebooks: Can they replace paper?” Journal of Cheminformatics, vol. 9, no. 1, p. 31, May 24, 2017, issn: 1758-2946. doi: http://doi.org/10.1186/s13321-017-0221-3. [Online]. Available: https://doi.org/10.1186/s13321-017-0221-3.
[25] S. G. Higgins, A. A. Nogiwa-Valdez, and M. M. Stevens, “Considerations for implementing electronic laboratory notebooks in an academic research environment,” Nature Protocols, vol. 17, no. 2, pp. 179–189, 2 Feb. 2022, issn: 1750-2799. doi: http://doi.org/10.1038/s41596-021-00645-8. [Online]. Available: https://www.nature.com/articles/s41596-021-00645-8 (visited on 01/20/2023).
[26] F. Abdullah, R. Ward, and E. Ahmed, “Investigating the influence of the most commonly used external variables of TAM on students’ Perceived Ease of Use (PEOU) and Perceived Usefulness (PU) of e-portfolios,” Computers in Human Behavior, vol. 63, no. C, pp. 75–90, Oct. 1, 2016, issn: 0747-5632. doi: http://doi.org/10.1016/j.chb.2016.05.014. [Online]. Available: https://doi.org/10.1016/j.chb.2016.05.014.
[27] Harvard Medical School, Boston, MA. “Electronic Lab Notebooks.” (2023), [Online]. Available: https://datamanagement.hms.harvard.edu/collect-analyze/electronic-lab-notebooks (visited on 09/27/2023).
[28] N. Carpi, A. Minges, and M. Piel, “eLabFTW: An open source laboratory notebook for research labs,” Journal of Open Source Software, vol. 2, no. 12, p. 146, Apr. 14, 2017, Sources: https://github.com/elabftw/, issn: 2475-9066. doi: http://doi.org/10.21105/joss.00146. [Online]. Available: https://joss.theoj.org/papers/10.21105/joss.00146.
[29] P. Tremouilhac, A. Nguyen, Y.-C. Huang, et al., “Chemotion ELN: An Open Source electronic lab notebook for chemists in academia,” Journal of Cheminformatics, vol. 9, no. 1, p. 54, Sep. 25, 2017, issn: 1758-2946. doi: http://doi.org/10.1186/s13321-017-0240-0. [Online]. Available: https://doi.org/10.1186/s13321-017-0240-0.
[30] RSpace ELN, Formerly eCAT from the University of Wisconsin., Edinburgh, Scotland: Research Space. [Online]. Available: https://www.researchspace.com/ (visited on 07/12/2023).
[31] eLabJournal, eLabNext. [Online]. Available: https://www.elabnext.com/products/elabjournal/.
[32] C. Draxl and M. Scheffler, “The NOMAD laboratory: From data sharing to artificial intelligence,” Journal of Physics: Materials, vol. 2, no. 3, p. 036 001, May 2019, Sources: https://gitlab.mpcdf.mpg.de/nomad-lab/nomad-FAIR, issn: 2515-7639. doi: http://doi.org/10.1088/2515-7639/ab13bb. [Online]. Available: https://dx.doi.org/10.1088/2515-7639/ab13bb.
[33] L. Patiny, M. Zasso, D. Kostro, et al., “The C6H6 NMR repository: An integral solution to control the flow of your data from the magnet to the public,” Magnetic Resonance in Chemistry, vol. 56, no. 6, pp. 520–528, 2018, issn: 1097-458X. doi: http://doi.org/10.1002/mrc.4669. [Online]. Available: https://onlinelibrary.wiley.com/doi/abs/10.1002/mrc.4669.
[34] T. Bronger. “Introduction — JuliaBase, the samples database.” (2023), [Online]. Available: https://www.juliabase.org/ (visited on 09/27/2023).
[35] G. King, “An Introduction to the Dataverse Network as an Infrastructure for Data Sharing,” Sociological Methods & Research, vol. 36, no. 2, pp. 173–199, Nov. 1, 2007, issn: 0049-1241. doi: http://doi.org/10.1177/0049124107306660. [Online]. Available: https://doi.org/10.1177/0049124107306660.
[36] J. Caffaro and S. Kaplun. “Invenio: A Modern Digital Library for Grey Literature.” Sources: https://github.com/inveniosoftware. (2010), [Online]. Available: https://cds.cern.ch/record/1312678 (visited on 07/12/2023), preprint.
[37] M. Smith, M. Barton, M. Bass, et al., “DSpace: An Open Source Dynamic Digital Repository,” Jan. 2003, issn: 1082-9873. doi: http://doi.org/10.1045/january2003-smith. [Online]. Available: https://dspace.mit.edu/handle/1721.1/29465 (visited on 07/12/2023).
[38] J. Winn, “Open data and the academy: An evaluation of CKAN for research data management,” presented at the IASSIST 2013, Cologne, May 2013. [Online]. Available: http://eprints.lincoln.ac.uk/id/eprint/9778/ (visited on 07/12/2023).
[39] A. Ailamaki, V. Kantere, and D. Dash, “Managing scientific data,” Communications of the ACM, vol. 53, no. 6, pp. 68–78, Jun. 1, 2010, issn: 0001-0782. doi: http://doi.org/10.1145/1743546.1743568. [Online]. Available: https://doi.org/10.1145/1743546.1743568.
[40] A. Nourani, H. Ayatollahi, and M. S. Dodaran, “Clinical Trial Data Management Software: A Review of the Technical Features,” Reviews on Recent Clinical Trials, vol. 14, no. 3, pp. 160–172, Sep. 1, 2019. doi: http://doi.org/10.2174/1574887114666190207151500.
[41] O. Hartig, P.-A. Champin, and G. Kellogg. “RDF 1.2 Concepts and Abstract Syntax.” (Jun. 29, 2023), [Online]. Available: https://www.w3.org/TR/2023/WD-rdf12-concepts-20230629/ (visited on 07/12/2023).
[42] S. Harris and A. Seaborne. “SPARQL 1.1 Query Language.” (Mar. 21, 2013), [Online]. Available: https://www.w3.org/TR/2013/REC-sparql11-query-20130321/ (visited on 07/12/2023).
[43] D. Vrandečić and M. Krötzsch, “Wikidata: A free collaborative knowledgebase,” Communications of the ACM, vol. 57, no. 10, pp. 78–85, Sep. 23, 2014, issn: 0001-0782, 1557-7317. doi: http://doi.org/10.1145/2629489. [Online]. Available: https://dl.acm.org/doi/10.1145/2629489.
[44] N. Brandt, L. Griem, C. Herrmann, et al., “Kadi4Mat: A Research Data Infrastructure for Materials Science,” vol. 20, no. 1, p. 8, 1 Feb. 10, 2021, issn: 1683-1470. doi: http://doi.org/10.5334/dsj-2021-008. [Online]. Available: https://datascience.codata.org/articles/10.5334/dsj-2021-008 (visited on 09/27/2023).
[45] T. P. Raptis, A. Passarella, and M. Conti, “Data Management in Industry 4.0: State of the Art and Open Challenges,” IEEE Access, vol. 7, pp. 97 052–97 093, Jul. 16, 2019, issn: 2169-3536. doi: http://doi.org/10.1109/ACCESS.2019.2929296.
[46] S. V. Tuyl and A. Whitmire, “Investigation of Non-Academic Data Management Practices to Inform Academic Research Data Management,” Research Ideas and Outcomes, vol. 4, e30829, Oct. 31, 2018, issn: 2367-7163. doi: http://doi.org/10.3897/rio.4.e30829. [Online]. Available: https://riojournal.com/article/30829/ (visited on 01/19/2023).
[47] M. A. Kennan and L. Markauskaite, “Research Data Management Practices: A Snapshot in Time,” International Journal of Digital Curation, vol. 10, no. 2, pp. 69–95, 2 Jun. 30, 2015, issn: 1746-8256. doi: http://doi.org/10.2218/ijdc.v10i2.329. [Online]. Available: http://ijdc.net/index.php/ijdc/article/view/10.2.69 (visited on 01/19/2023).
[48] LinkAhead. “HUMANS.md,” GitLab. (Sep. 13, 2023), [Online]. Available: https://gitlab.com/linkahead/linkahead/-/blob/main/HUMANS.md (visited on 10/05/2023).
[49] “Tutorial: Parameter file — LinkAhead Crawler 0.7.0 documentation.” (2023), [Online]. Available: https://docs.indiscale.com/caosdb-crawler/tutorials/parameterfile.html#getting-started-with-the-cfood (visited on 07/12/2023).
[50] “R2RML: RDB to RDF Mapping Language.” (Sep. 27, 2012), [Online]. Available: https://www.w3.org/TR/2012/REC-r2rml-20120927/ (visited on 07/12/2023).
[51] F. Spreckelsen, B. Rüchardt, J. Lebert, S. Luther, U. Parlitz, and A. Schlemmer, “Guidelines for a Standardized Filesystem Layout for Scientific Data,” Data, vol. 5, no. 2, p. 43, 2 Jun. 2020, issn: 2306-5729. doi: http://doi.org/10.3390/data5020043. [Online]. Available: https://www.mdpi.com/2306-5729/5/2/43 (visited on 01/18/2023).
[52] “LinkAhead / LinkAhead Crawler Cfoods / ELabFTW Cfood · GitLab,” GitLab. (Oct. 5, 2023), [Online]. Available: https://gitlab.com/linkahead/crawler-extensions/elabftw-cfood (visited on 10/05/2023).
[53] “LinkAhead Query Language Examples — linkahead-server 0.11.0 documentation.” (2023), [Online]. Available: https://docs.indiscale.com/caosdb-server/LinkAhead-Query-Language.html (visited on 10/05/2023).
[54] “#Linkahead on Matrix chat.” (Oct. 5, 2023), [Online]. Available: https://matrix.to/#/#linkahead:matrix.org (visited on 10/05/2023).
[55] “The ELN Consortium,” GitHub. (2023), [Online]. Available: https://github.com/TheELNConsortium (visited on 07/12/2023).
[56] P. Sefton, E. Ó Carragáin, S. Soiland-Reyes, et al., “RO-Crate Metadata Specification 1.1.3,” Apr. 26, 2023. doi: http://doi.org/10.5281/zenodo.7867028. [Online]. Available: https://zenodo.org/record/7867028 (visited on 07/12/2023).
[57] S. Soiland-Reyes, P. Sefton, M. Crosas, et al., “Packaging research artefacts with ROCrate,” Data Science, vol. 5, no. 2, pp. 97–138, Jan. 1, 2022, issn: 2451-8484. doi: http://doi.org/10.3233/DS-210053. [Online]. Available: https://content.iospress.com/articles/data-science/ds210053 (visited on 07/12/2023).
[58] “LinkAhead Enhancement Requests,” GitLab - LinkAhead. (Oct. 5, 2023), [Online]. Available: https://gitlab.com/groups/linkahead/-/issues?or[label_name][]=Enhancement%3A%3AAccepted&or[label_name][]=Enhancement%3A%3AProposed&state=opened (visited on 10/05/2023).
[59] “LinkAhead Trino branch,” GitLab LinkAhead. (Oct. 5, 2023), [Online]. Available: https://gitlab.com/linkahead/linkahead-server/-/tree/f-experiment-trino (visited on 10/05/2023).