1 Introduction
With increasing volume, complexity and creation speed of scholarly data, humans rely more and more on computational support in processing this data. The “FAIR guiding principles for scientific data management and stewardship” [1] were introduced in order to improve the ability of machines to automatically find and use that data. FAIR comprises the four foundational principles “that all research objects should be Findable, Accessible, Interoperable and Reusable (FAIR) both for machines and for people”. In giving abstract, high-level and domain-independent guidelines, the authors answer the question of what constitutes good data management. However, the implementation of these guidelines is still in its infancy, with many challenges not yet identified and some of which may not have readily available solutions [2]. Furthermore, efforts are made towards an Internet of FAIR Data and Services (IFDS) [3], which requires not only the data, but also the tools and (compute) services to be FAIR.
Data processing is usually not a single task, but in general (and in particular for computational simulations) relies on a chain of tools. Thus, to achieve transparency, adaptability and reproducibility of (computational) research, the FAIR principles must be applied to all components of the research process. This includes the tools (i. e. any research software) used to analyze the data, but also the scientific workflow itself which describes how the various processes depend on each other. In a community-driven effort, the FAIR principles were applied to research software and extended to its specific characteristics by the FAIR for Research Software Working Group [4]. For a discussion of how the FAIR principles should apply to workflows and workflow management systems (WfMSs) we refer to [5].
In addition, in recent years there has been a tremendous development of different tools (see e.g. https://github.com/pditommaso/awesome-pipeline) that aid the definition and automation of computational workflows. These WfMSs have great potential in supporting the goal above which is further discussed in section 1.1. The key features of WfMSs are also highlighted in the context of bioinformatics workflows by [6], which compare several WfMSs regarding aspects ranging from portability over scalability to the availability of learning resources. A discussion of strengths and weaknesses of a selection of tools in the context of material sciences is given in a recent work [7].
In this work, we would like to discuss how WfMSs can contribute to the transparency, adaptability, reproducibility and reusability of computational research. Similar to [6], [7], we evaluate a selection of WfMSs regarding a set of requirements, taking into account different possible scenarios in which WfMSs are employed. In contrast to [7], we consider generic scenarios that are not tied to a specific research domain (see section 2), from which we derive requirements on WfMSs that we deem relevant in those contexts (see section 3). This leads to a set of requirements that overlap with the ones presented in [6], but include more specific aspects of workflow definitions and their development process. While [4], [5] discuss properties of FAIR research software and workflows on a rather high level, this work focuses on how concrete features of WfMSs may contribute to a more FAIR research software landscape. However, with the considered requirements, we focus on the aspects reusability and interoperability, since findability and accessibility lie outside the responsibilities of a WfMS.
Several WfMSs are evaluated with respect to the requirements by means of an exemplary workflow, which is described in section 4, in addition to the available online documentation (see below). The evaluation is presented in section 5, with the aim to support fellow scientists in identifying the tools that best suit their requirements. The list of tools selected for comparison is subjective and certainly not complete. However, a GitHub repository [8] providing an implementation of the exemplary workflow for all tools and a short documentation with a link to further information was created, with the aim to continuously add more tools in the future. Furthermore, by demonstrating how the different tools could be used, we would like to encourage people to use WfMSs in their daily work.
1.1 Introduction to workflow management systems
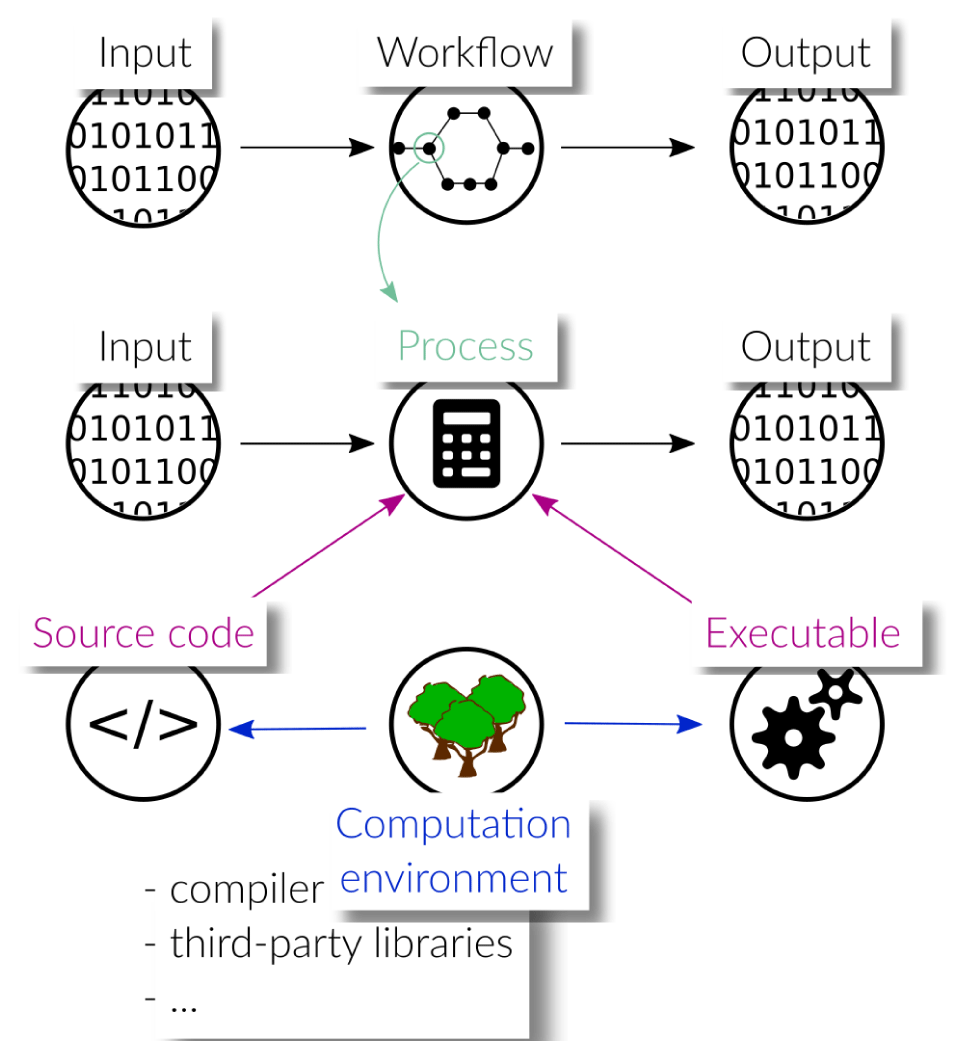
In this paper, we use the term process to describe a computation, that is, the execution of a program to produce output data from input data. A process can be arbitrarily complex, but from the point of view of the workflow, it is a single, indivisible step. A workflow describes how individual processes relate to each other. Software-driven scientific workflows are often characterized by a complex interplay of various pieces of software executed in a particular order. The output of one process may serve as input to a subsequent process, which requires them to be executed sequentially with a proper mapping of outputs to inputs. Other computations are independent of each other and can be executed in parallel. Thus, one of the main tasks of WfMSs is the proper and efficient scheduling of the individual processes.
As shown in fig. 1, each process in the workflow, just as the workflow itself, takes some input to produce output data. A more detailed discussion of the different levels of abstractions related to workflows can be found in Griem et al. [9]. The behavior of a process is primarily determined by the source code that describes it, but may also be influenced by the interpreters/compilers used for translation or the machines used for execution. Moreover, the source code of a process may carry dependencies to other software packages such that the behavior of a process possibly depends on their versions. We use the term computation environment to collect all those software dependencies, that is, interpreters and/or compilers as well as third-party libraries and packages that contribute to the computations carried out in a process. The exact version numbers of all involved packages are crucial, as the workflow may not work with newer or older packages, or, may produce different results.
As outlined in [10], WfMSs may be grouped into five classes. First, tools like Galaxy [11], KNIME [12], and Pegasus [13] provide a graphical user interface (GUI) to define scientific workflows. Thus, no programming skills are required and the WfMS is easily accessible for everybody. With the second group, workflows are defined using a set of classes and functions for generic programming languages (libraries and packages). This has the advantage that version control (e.g. using Git (https://git-scm.com)) can be employed on the workflow. In addition, the tool can be used without a graphical interface, e.g. in a server environment. Examples of prominent tools are AiiDA [14], [15], doit [16], Balsam [17], FireWorks [18], SciPipe [19] and Guix Workflow Language [20]. Third, tools like Nextflow [21], Snakemake [22], Bpipe [23] and Cluster Flow [24] express the workflow using a domain specific language (DSL). A DSL is a language tailored to a specific problem. In this context, it offers declarations and statements to implement often occurring constructs in workflow definitions, which improves the readability and reduces the amount of code. Moreover, the advantages of the second group also apply for the third group. In contrast to the definition of the workflow in a programmatic way, the fourth group comprises tools like Popper [25] and Argo workflows (https://argoproj.github.io/argo-workflows/) which allow to specify the workflow in a purely declarative way, by using configuration file formats like YAML [26]. In this case, the workflow specification is concise and can be easily understood, but lacks expressiveness compared to the definition of the workflow using programming languages. Fifth, there are system-independent workflow specification languages like CWL [27] or WDL (https://github.com/openwdl/wdl). These define a declarative language standard for describing workflows, which can then be executed by a number of different engines like Cromwell [28], Toil [29], and Tibanna [30].
WfMSs can be used to create, execute and monitor workflows. They can help to achieve reproducibility of research results by avoiding manual steps and automating the execution of the individual processes in the correct order. More importantly, for a third person to reproduce and reuse the workflow, it needs to be portable, that is, executable on any machine. Portability can be supported by WfMSs with the integration of package management systems and container technologies, which allow them to automatically re-instantiate the compute environment. Another advantage of using WfMSs is the increase in transparency through a clear and readable workflow specification. Moreover, after completion of the workflow, the tool can help to trace back a computed value to its origin, by logging all inputs, outputs and possibly metadata of all computations.
2 User stories
Starting from user stories that we consider representative for computational science and engineering, a set of requirements is derived that serves as a basis for the comparison of different WfMSs. In particular, a discussion on how the different tools implement these requirements is provided.
Reproducibility, which is key to transparent research, is the main focus of the first user story (see section 2.1). The second user story (see section 2.2) deals with research groups that develop workflows in a joint effort where subgroups or individuals work on different components of the workflow. Finally, the third user story focuses on computational research that involves generating and processing large amounts of data, which poses special demands on how the workflow tools organize the data that is created upon workflow execution (see section 2.3).
2.1 Transparent and reproducible research paper
As a researcher, I want to share the code for my paper such that others are able to easily reproduce my results.
In this user story, the main objective is to guarantee the reproducibility of computational results presented in scientific publications. Here, reproducibility means that a peer researcher is able to rerun the workflow on some other machine while obtaining results that are in good agreement with those reported in the publication. Mere reproduction could also be achieved without a WfMS, e.g. by providing a script that executes the required commands in the right order, but this comes with a number of issues that may be solved with a standardized workflow description.
First of all, reconstructing the logic behind the generation and processing of results directly from script code is cumbersome and reduces the transparency of the research, especially for complex workflows. Second, it is not straightforward for peer researchers to extract certain processes of a workflow from a script and embed them into a different research project, hence the reusability aspect is poorly addressed with this solution. Workflow descriptions may provide a remedy to both of these issues, provided that each process in the workflow is defined as a unit with a clear interface (see section 3.7).
While the workflow description helps peers to understand the details behind a research project, it comes with an overhead on the side of the workflow creator, in particular when using a WfMS for the first time. In the prevalent academic climate, but also in industrial (research) settings, we therefore think that an important aspect of WfMSs is how easy they are to get started with (see section 3.9). Similarly, if the WfMS provides a GUI to visualize and/or define the workflow (see section 3.3), no special programming skills are required, which may be preferred due to the easy access.
In the development phase, a workflow is typically run many times until its implementation is satisfactory. With a scripted automation, the entire workflow is always executed, even if only one process was changed since the last run. Since WfMSs have to know the dependencies between processes, this opens up the possibility to identify and select only those parts of a workflow that have to be rerun (see section 3.8). Besides this, the WfMS can display to the user which parts are currently being executed, which ones have already been up-to-date, and which ones are still to be picked (see section 3.2).
A general issue is that a workflow, or even each process in it, has a specific set of software- and possibly hardware-requirements. This makes both reproducibility and reusability difficult to achieve, especially over longer time scales, unless the computation environment in which the original study was carried out is documented in a way that allows for a later re-instantiation. The use of package managers that can export a given environment into a machine-readable format from which they can then recreate that environment at a later time, may help to overcome this issue. Another promising approach is to rely on container technologies. WfMSs have the potential to automate the re-instantiation of a computation environment via integration of either one of the above-mentioned technologies (see section 3.5). This makes it much easier for peers to reproduce and/or reuse parts of a published workflow.
2.2 Joint research (group)
As part of a research group, I want to be able to interconnect and reuse components of several different workflows so that everyone may benefit from their colleagues’ work.
Similar to the previous user story, the output of such a workflow could be a scientific paper. However, this user story explicitly considers interdisciplinary workflows in which the reusability of individual components/modules is essential. Each process in the workflow may require a different expertise and hence modularity and a common framework is necessary for an efficient collaboration.
Many of the difficulties discussed in the previous user story are shared in a joint research project. However, the collaborative effort in which the workflow description and those of its components are developed promotes the importance of clear interfaces (see section 3.7) to ease communication and an intuitive dependency handling mechanism (see section 3.5).
As mentioned in section 2.1, a GUI can help to increase the usability of a workflow for non-programmers. However, in this user story it is important that the workflow definition is available in a human-readable and manually editable format (see section 3.10). This facilitates version control and the code review process as an essential aspect of teamwork.
Another challenge here is that such workflows often consist of heterogeneous models of different complexity, such as large computations requiring high-performance computing (HPC), preprocessing of experimental data or postprocessing analyses. Due to this heterogeneity, it may be beneficial to outsource computationally demanding tasks to HPC systems, while executing cheaper tasks locally (see section 3.1). Workflows with such computationally expensive tasks can also strongly benefit from effective caching mechanisms and the reuse of cached results wherever possible (see section 3.8).
Finally, support for a hierarchical embedding of sub-workflows (possibly published and versioned) in another workflow is of great benefit as this allows for an easy integration of improvements made in the sub-workflows by other developers (see section 3.6).
2.3 Complex hierarchical computations
As a materials scientist, I want to be able to automate and manage complex workflows so I can keep track of all associated data.
Workflows in which screening or parameter sweeps are required typically involve running a large number of simulations. Moreover, these workflows are often very complex with many levels of dependencies between the individual tasks. Good data management that provides access to the full provenance graph of all data can help to retain an overview over the large amounts of data produced by such workflows (see section 3.4). For instance, the data management could be such that desired information may be efficiently extracted via query mechanisms.
Another aspect regarding high-throughput computational screening is that the same computations are carried out for many inputs (material structures) and the same workflow might be used for a number of studies on varying input data. Here, a platform for publishing and sharing workflows (see section 3.11) with the community can help to standardize and assure the quality of the workflow. Furthermore, the findability and accessibility of workflows are increased, thereby contributing to open science.
Due to the large amount of computationally demanding tasks in such workflows, it is helpful if some computations can be outsourced to HPC systems (see section 3.1) with a clean way of querying the current status during the typically long execution times (see section 3.2).
3 Specific requirements on workflow management systems
The user stories described above allow us to identify 11 requirements on WfMSs. Some of these requirements concern the interaction with a WfMS from the perspective of a user of a workflow, while others are related to the creation of a workflow definition and its readability or portability. While portability is key to reproducible research, readability is an important aspect of transparency. However, an easy and intuitive way of interacting with a WfMS is crucial for workflows to be reused at all. Finally, reusability is enhanced if the workflow, or parts of it, can be embedded into another workflow in a possibly different context. In the following, we will describe the requirements in detail, as they will serve as evaluation criteria for the individual WfMSs discussed in section 5.
3.1 Support for job scheduling system
As already mentioned, the main task of a WfMS is to automatically execute the processes of a workflow in the correct order such that the dependencies between them are satisfied. However, processes that do not depend on each other may be executed in parallel in order to speed up the overall computation time. This requirement focuses on the ability of a WfMS to distribute the computations on available resources. Job scheduling systems like e.g. Slurm (also commonly referred to as batch scheduling or batch systems) are often used to manage computations to be run and their resource requirements (number of nodes, CPUs, memory, runtime, etc.). Therefore, it is of great benefit if WfMSs support the integration of widely-used batch systems such that users can specify and also observe the used resources alongside other computations that were submitted to their batch system in use. Besides this, this requirement captures the ability of a WfMS to outsource computations to a remote machine, e.g. a HPC cluster or cloud. In this sense, this requirement is crucial for workflows that require HPC resources to be reproducible. For traditional HPC cluster systems it is usually necessary to transfer input and output data between the local system and the cluster system. This can be done using the secure shell protocol (SSH) and a WfMS may provide the automated transfer of a job’s associated data. Ideally, the workflow can be executed anywhere without changing the workflow definition itself, but only the runtime arguments or a configuration file. The fulfillment of this requirement is evaluated by the following criteria:
The workflow system supports the execution of the workflow on the local system.
The workflow system supports the execution of the workflow on the local system via a batch system.
The workflow system supports the execution of the workflow via a batch system on the local or a remote system.
3.2 Monitoring
Depending on the application, the execution of scientific workflows can be very time-consuming. This can be caused by compute-intensive processes such as numerical simulations, or by a large number of short processes that are executed many times. In both cases, it can be very helpful to be able to query the state of the execution, that is, which processes have been finished, which processes are currently being executed, and which are still pending. A trivial way of such monitoring would be, for instance, when the workflow is started in a terminal which is kept open to inspect the output written by the workflow system and the running processes. However, ideally, the workflow system allows for submission of the workflow in the form of a process running in the background, while still providing means to monitor the state of the execution. For this requirement, two criteria are distinguished:
The only way to monitor the workflow is to watch the console output.
The workflow system provides a way to query the execution status at any time.
3.3 Graphical user interface
Independent of a particular execution of the workflow, the workflow system may provide facilities to visualize the graph of the workflow, indicating the mutual dependencies of the individual processes and the direction of the flow of data. One can think of this graph as the template for the data provenance graph. This visualization can help in conveying the logic behind a particular workflow, making it easier for other researchers to understand and possibly incorporate it into their own research. The latter requires that the workflow system is able to handle hierarchical workflows, that is, workflows that contain one or more sub-workflows as processes (see section 3.6). Beyond a mere visualization, a GUI may allow for visually connecting different workflows into a new one by means of drag & drop. We evaluate the features of a graphical user interface by means of the following three criteria:
The workflow system provides no means to visualize the workflow
The workflow system or third-party tools allow to visualize the workflow definition
The workflow system or third-party tools provide a GUI that enables users to graphically create workflows
3.4 Data provenance
The data provenance graph contains, for a particular execution of the workflow, which data and processes participated in the generation of a particular piece of data. Thus, this is closely related to the workflow itself, which can be thought of as a template for how that data generation should take place. However, a concrete realization of the workflow must contain information on the exact input data, parameters and intermediate results, possibly along with meta information on the person that executed the workflow, the involved software, the compute resources used and the time it took to finish. Collection of all relevant information, its storage in machine-readable formats and subsequent publication alongside the data can be very useful for future researchers in order to understand how exactly the data was produced, thereby increasing the transparency of the workflow and the produced data. Ideally, the workflow system has the means to automatically collect this information upon workflow execution, which we evaluate using the following criteria:
The workflow system provides no means to export relevant information from a particular execution
The workflow system stores all results (also intermediate) together with provenance metadata about how they were produced
3.5 Compute environment
In order to guarantee interoperability and reproducibility of scientific workflows, the workflows need to be executable by others. Here, the re-instantiation of the compute environment (installation of libraries or source code) poses the main challenge. Therefore, it is of great use if the WfMS is able to automatically deploy the software stack (on a per workflow or per process basis) by means of a package manager (e.g. conda https://conda.io/) or that running processes in a container (e.g. Docker https://www.docker.com, Apptainer https://apptainer.org (formerly Singularity)) is integrated in the tool. The automatic deployment of the software stack facilitates the execution of the workflow, and thus, greatly enhances its reproducibility. However, it does not (always) enable reusage, that is, the associated software can be understood, modified, built upon or incorporated into other software [4]. For instance, if a container image is used, it is important that the container build recipe (e.g. Dockerfile) is provided. This increases the reusability as it documents how a productive environment, suitable to execute the given workflow or process, can be set up. The author of the workflow, however, is deemed to be responsible for the documentation of the compute environment. For this requirement, we define the following evaluation criteria:
The automatic instantiation of the compute environment is not intended.
The workflow system allows the automatic instantiation of the compute environment on a per workflow basis.
The workflow system allows the automatic instantiation of the compute environment on a per process basis.
3.6 Hierarchical composition of workflows
A workflow consists of a mapping between a set of inputs (could be empty) and a set of outputs, whereas in between a number of processes are performed. Connecting the output of one workflow to the input of another workflow results in a new, longer workflow. This is particularly relevant in situations where multiple people share a common set of procedures (e.g. common pre- and postprocessing routines). In this case, copying the preprocessing workflow into another one is certainly always possible, but does not allow to jointly perform modifications and work with different versions. Moreover, a composition might also require to define separate compute environments for each sub-workflow (e.g. using Docker/singularity or conda). Executing all sub-workflows in the same environment might not be possible because each sub-workflow might use different tools or even the same tools but with different versions (e.g. python2 vs. python3). Thus, WfMSs that can incorporate other workflows, possibly executed in a different compute environment, increase the reusability of a workflow substantially. This promotes the importance of supporting heterogeneous compute environments, which is reflected in the evaluation criteria for this requirement:
The workflow system does not allow the composition of workflows.
The workflow system allows to embed a workflow into another one for a single compute environment (homogeneous composition).
The workflow system allows to embed a workflow into another one for arbitrary many (on a per process basis) compute environments (hierarchical composition).
3.7 Interfaces
In a traditional file-based pipeline, the output files produced by one process are used as inputs to a subsequent process. However, it is often more convenient to pass non-file output (e.g. float or integer values) directly from one process to another without the creation of intermediate files. In this case, it is desirable that the WfMS is able to check the validity of the data (e.g. the correct data type) to be processed. Furthermore, this defines the interface for a process more clearly and makes it easier for someone else to understand how to use, adapt or extend the workflow/process. In contrast, in a file-based pipeline, this is usually not the case since a dependency in form of a file does not give information about the type of data contained in that file. For the sake of transparency and reusability, it is beneficial if a WfMS supports the definition of strongly-typed process interfaces. Type-checking the workflow definition before execution can also help to avoid unnecessary computations with erroneous workflows that attempt to transfer data with incompatible types. We distinguish these different types of interfaces by the following criteria:
The workflow system is purely file-based and does not define interface formats.
The workflow system allows for passing file and non-file arguments between processes.
The workflow system allows for defining strongly-typed process interfaces, supporting both file and non-file arguments.
3.8 Up-to-dateness
There are different areas for the application of workflows. On the one hand, people might use a workflow to define a single piece of reproducible code that, when executed, always returns the same result. Based on that, they might start a large quantity of different jobs and use the workflow system to perform this task. Another area of application is the constant development within the workflow (e.g. exchanging processes, varying parameters or even modifying the source code of a process) until a satisfactory result is obtained. The two scenarios require a slightly different behavior of the workflow system. In the first scenario, all runs should be kept in the data provenance graph with a documentation of how each result instance has been obtained (e.g. by always documenting the codes, parameters, and processes). If identical runs (identical inputs and processes should result in the same output) are detected, a recomputation should be avoided and the original output should be linked in the data provenance graph. The benefit of this behavior certainly depends on the ratio between the computation time for a single process compared to the overhead to query the data base.
However, when changing the processes (e.g. coding a new time integration scheme or a new constitutive model), the workflow system should rather behave like a build system (such as make) - only recomputing the steps that are changed or that depend on these changes. In particular for complex problems, this allows to work with complex dependencies without manually triggering computations and results in automatically recomputing only the relevant parts. An example is a paper with multiple figures where each is a result of complex simulations that in itself depend on a set of general modules developed in the paper. The “erroneous” runs are usually not interesting and should be overwritten.
How this is handled varies between the tools, yielding the following evaluation criteria:
R The complete workflow is always Recomputed.
L A new entry in the data provenance graph is created which Links the previous result (without the need to recompute already existing results).
U Only the parts are recreated (Updated) that are not up-to-date. This usually reduces the overhead to store multiple instances of the workflow, but at the same time also prevents - without additional effort (e.g. when executing in different folders) computing multiple instances of the same workflow.
3.9 Ease of first use
Although this is not a requirement per-se, it is beneficial if the workflow system has an intuitive syntax/interface and little work is required for a new user to define a first workflow. Research applications typically have a high intrinsic complexity, and therefore, the complexity added by the workflow management should be as small as possible. We note that this requirement is subjective and depends on the experience and skills of the user. Nevertheless, from the perspective of engineers and self-taught programmers, the following criteria are defined, considering aspects such as readability, expressiveness and knowledge of the tool:
difficult: Extensive knowledge of the tool and its design concepts as well as advanced programming skills are required to define a first workflow.
intermediate: Extensive knowledge of the tool and its design concepts and only basic programming skills are required to define a first workflow.
easy: Only basic programming skills are required to define a first workflow.
3.10 Manually editable workflow definition
While it can be beneficial to create and edit workflows using a GUI (see section 3.3), it may be important that the resulting workflow description is given in a human-readable format. This does not solely mean that the definition should be a text file, but also that the structure (e.g. indentation) and the naming are comprehensive. This facilitates version-controlling with git, and in particular the code review process. This increases the transparency of a workflow, and moreover, this does not force all users and/or developers to rely on the GUI. Evaluation criteria:
The workflow description is a binary file.
The workflow description is a text file but hard to interpret by humans.
The workflow description is a fully human-readable file format.
3.11 Platform for publishing and sharing workflows
The benefit of a workflow system is already significant when using it for individual research such as the development of an individual’s paper or reproducing the paper that someone else has written, when their data processing pipeline is fully reproducible, documented and published. However, the benefit can be even more increased if people are able to jointly work on (sub-)workflows together; particularly when a hierarchical workflow system is used. Even though workflows can easily be shared together with the work (e.g. in a repository), it might be beneficial to provide a platform that allows to publish documented workflows with a search and versioning functionality. This feature is not part of the requirement matrix to compare the different tools, but we consider a documentation of these platforms in the subsequent section as a good starting point for further research (exchange).
4 Exemplary workflow
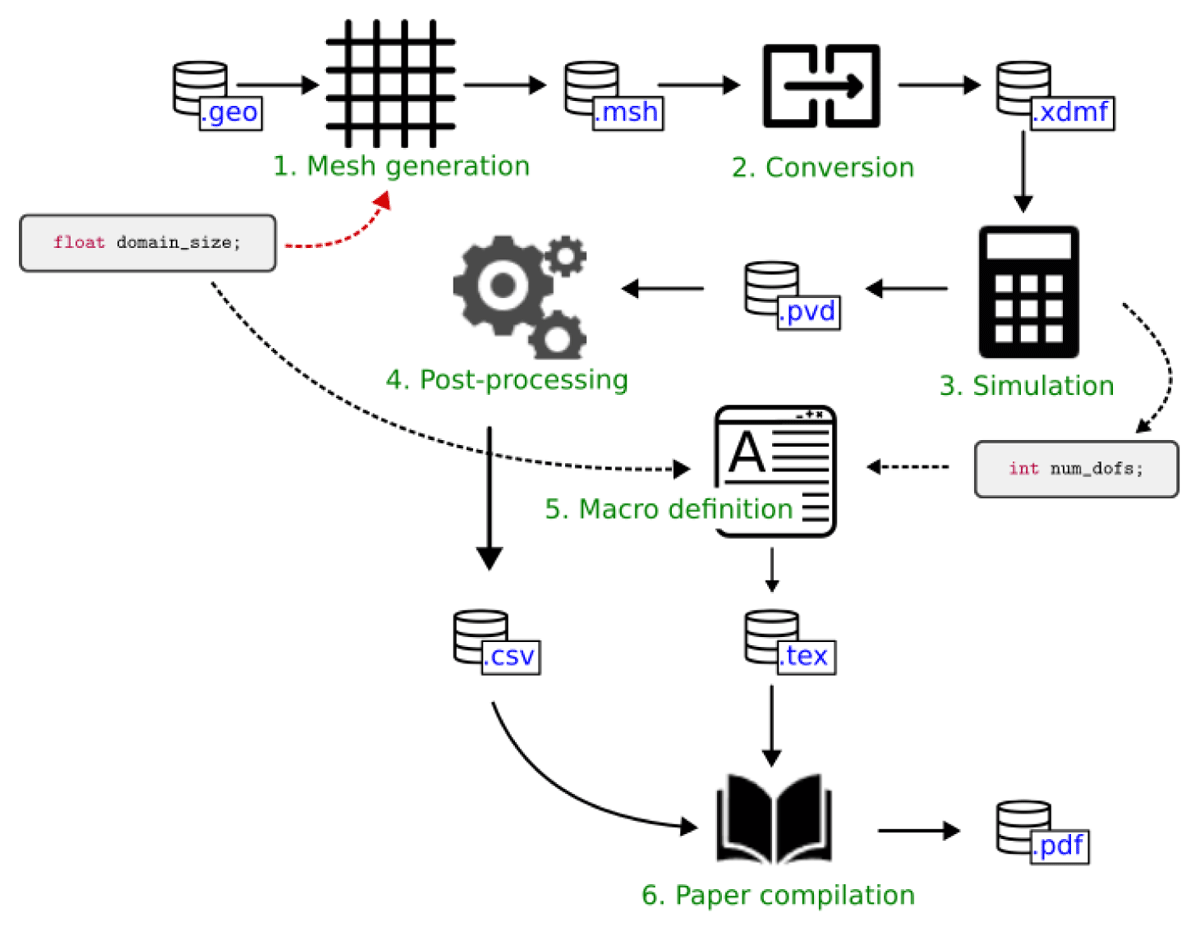
A simple exemplary workflow was defined in order to analyze and evaluate the different WfMSs with respect to the requirements stated in section 3. This example is considered to be representative for many problems simulating physical processes in engineering science using numerical discretization techniques. It consists of six steps, as shown in fig. 2:
generation of a computational mesh (Gmsh)
mesh format conversion (MeshIO)
numerical simulation (FEniCS)
post-processing of the simulation results (ParaView)
preparation of macro definitions (Python)
compilation of a paper into a .pdf file using the simulation results (Tectonic)
The workflow starts from a given geometry on which the simulation should be carried out and generates a computational mesh in the first step using Gmsh [31]. Here, the user can specify the size of the computational domain by a float value domain_size. The resulting mesh file format is not supported by FEniCS [32], which is the software that we are using for the simulation carried out in the third step. Therefore, we convert the mesh file in the second step of the workflow from .msh to .xdmf using the python package MeshIO [33]. The simulation step yields result files in VTK file format [34] and returns the number of degrees of freedom used by the simulation as an integer value num_dofs. The VTK files are further processed using the python application programming interface (API) of ParaView [35], which yields the data of a plot-over-line of the numerical solution across the domain in .csv file format. This data, together with the values for the domain size and the number of degrees of freedom, is inserted into the paper and compiled into a .pdf file using the LATEX engine Tectonic [36] in the final step of the workflow.
Most steps transfer data among each other via files, but we intentionally built in the transfer of the number of degrees of freedom as an integer value to check how well such a situation can be handled by the tools. Example implementations of the exemplary workflow for various tools are available in a public repository [8].
5 Tool comparison
In this section, the selected WfMSs and their most important features are described and set in relation to the requirements defined in section 3. We note that to the best of our knowledge, existing add-on packages to the individual WfMSs are as well considered. As mentioned in the introduction, a large number of WfMSs exist, and the ones selected in this work represent only a small fraction of them. The considered WfMSs were selected on the basis of their popularity within the authors’ communities, however, this has no implications on the quality of WfMSs not considered in this work. As mentioned, we also plan to include implementations of the exemplary workflow with further WfMSs in the online documentation in the future.
5.1 AiiDA
AiiDA [14], [15], the automated interactive infrastructure and database for computational science, is an open source Python infrastructure. With AiiDA, workflows are written in the Python programming language and managed and executed using the associated command line interface “verdi”.
AiiDA was designed for use cases that are more focused on running heavy simulation codes on heterogeneous compute hardware. Therefore, one of the key features of AiiDA is the HPC interface. It supports the execution of (sub-) workflows on any machine and most resource managers are integrated. In case of remote computing resources, any data transfer, retrieval and storing of the results in the database or status checking is handled by the AiiDA daemon process. Another key feature is AiiDA’s workflow writing system which provides strongly typed interfaces and allows for easy composition and reuse of workflows. Moreover, AiiDA automatically keeps track of all inputs, outputs and metadata of all calculations, which can be exported in the form of provenance graphs.
AiiDA’s workflow system enables to easily compose workflows, but a general challenge seems to be the management of the compute environment by the user. For external codes that do not run natively in Python the implementation of so-called plugins is required. The plugin instructs AiiDA how to run that code and might also contain (among other things) new data types or parsers that are necessary, for instance, to validate the calculated results before storing them in the database. Maintaining the plugin poses an additional overhead if the application code changes frequently during development of the workflow. Moreover, the user has to take care of the installation of the external code on the target computer.
However, since AiiDA version 2.1 it is possible to run code inside containers together with any existing plugin for that code. This mitigates the issue of the manual installation of the external code, but still requires a suitable plugin. Another benefit is that the information about the compute environment is stored in the database as well. At the time of writing, the containerization technologies Docker, Singularity https://singularity-docs.readthedocs.io/en/latest/ and Sarus https://sarus.readthedocs.io/en/stable/ are supported.
In the special case of FEniCS (see section 4), which can be used to solve partial differential equations and therefore covers a wide spectrum of applications, it is very difficult to define a general plugin interface which covers all models. We note that due to this use case, which is rather different from the use cases that AiiDA was designed for, the implementation of the exemplary workflow (see section 4) uses “aiida-shell” [37], an extension to the AiiDA core package which makes running shell commands easy. While this is convenient to get a workflow running quickly, this leads to an undefined process interface since this was the purpose of the aforementioned plugin for an external code. Considering the points above, compared to the other tools, the learning curve with AiiDA is fairly steep.
In contrast to file-based workflow management systems, AiiDA defines data types for any data that should be stored in a database. Consequently, non-file based inputs are well defined, but this is not necessarily the case for file data. The reason for the choice of a database is that it allows to query all stored data, and thus, enables powerful data analyses. For file-based workflows this is difficult to reproduce, especially for large amounts of data.
In terms of the requirements defined in section 3, AiiDA’s strong points are execution, monitoring and provenance. Due to the possibility to export provenance graphs, also level two of the requirement graphical user interface is reached. Lastly, caching can be enabled in AiiDA to save computation time. Caching in AiiDA means, that the database will be searched for a calculation of the same hash and if this is the case, the same outputs are reused.
5.2 Common Workflow Language
“Common Workflow Language (CWL) [38] is an open standard for describing how to run command line tools and connect them to create workflows” (https://www.commonwl.org/). One benefit of it being a standard is that workflows expressed in CWL do not have to be executed by a particular workflow engine, but can be run by any engine that is able to support the CWL standard. In fact, there exist a number of workflow engines that support CWL workflows, e.g. the reference implementation cwltool (https://github.com/common-workflow-language/cwltool), Toil [29] or StreamFlow [39]. Note that so far we have tested our implementation only with cwltool, however, in the evaluation we include all engines that support the CWL standard. That is, in this work we consider that CWL fulfills a specific requirement if there exists an engine that fulfills the requirement upon execution of a workflow written in CWL.
CWL was designed with a focus on data analysis using command line programs. To create a workflow, each of the command line programs is “wrapped” in a CWL description, defining what inputs are needed, what outputs are produced and how to call the underlying program. Typically, this step also reduces the possibly large number of options of the underlying command line tool to a few options or inputs that are relevant for the particular task of the workflow. In a workflow, the wrapped command line tools can be defined as individual processes, and the outputs of one process can be mapped to the inputs of other processes. This information is enough for the interpreter to build up the dependency graph, and processes that do not depend on each other may be executed in parallel. A process can also be another workflow, thus, hierarchical workflow composition is possible. Moreover, there exist workflow engines (e.g. Toil [29] or StreamFlow [39]) for CWL that support using job managers, for instance, Slurm [40].
The CWL standard also provides means to specify the software requirements of a process. For instance, one can provide the URL of a Docker image or Docker file to be used for the execution of a process. In case of the latter, the image is automatically built from the provided Docker file, which itself contains the information on all required software dependencies. Besides this, the CWL standard contains language features that allow listing software dependencies directly in the description of a workflow or process, and workflow engines may automatically make these software packages available upon execution. As one example, the current release of cwltool supports the definition of software requirements in the form of e.g. Conda packages that are then automatically installed when the workflow is run (see e.g. our implementation and the respective pipelines at [8]).
In contrast to workflow engines that operate within a particular programming language, the transfer of data from one process to another cannot occur directly via memory with CWL. For instance, if the result of a process is an integer value, this value has to be read from a file produced by the process, or, from its console output. However, this does not have to be done in a separate process or by again wrapping the command line tool inside some script, since CWL supports the definition of inline JavaScript code that is executed by the interpreter. This allows retrieving, for instance, integer or floating point return values from a process with a small piece of code.
CWL requires the types of all inputs and outputs to be specified, which has the benefit that the interpreter can do type checks before the execution of the workflow. A variety of primitive types, as well as arrays, files or directories, are available. Files can refer to local as well as online resources, and in the case of the latter, resources are automatically fetched and used upon workflow execution.
There exist a variety of tools built around the CWL standard, such as the Rabix Composer (https://rabix.io/) for visualizing and composing workflows in a GUI. Besides that and as mentioned before, there are several workflow engines that support CWL and some of which provide extra features. For instance, cwltool allows for tracking provenance information of individual workflow runs. However, to the best of our knowledge, there exists no tool that automatically checks which results are up-to-date and do not have to be reproduced (see section 3.8).
The CWL standard allows to specify the format of an input or output file by means of an IRI (Internationalized Resource Identifier) that points to online-available resource where the file format is defined. For processes whose output files are passed to the inputs of subsequent jobs, the workflow engine can use this information to check if the formats match. To the best of our knowledge, cwltool does so by verifying that the IRIs are identical, or performs further reasoning in case the IRIs point to classes in ontologies (see, for instance, the class for the JSON file format in the EDAM ontology at edamontology.org/format_3464). Such reasoning can make use of defined relationships between classes of the ontology to determine file format compatibility and thereby contribute to the requirement process interfaces. For more information on file format specifications in CWL see commonwl.org/user_guide/topics/file-formats.html.
5.3 doit
“doit comes from the idea of bringing the power of build-tools to execute any kind of task” [16]. The automation tool doit is written in the Python programming language. In contrast to systems which offer a GUI, knowledge of the programming language is required. However, it is not required to learn an additional API since task metadata is returned as a Python dictionary. Therefore, we consider this as very easy to get started quickly.
With doit, any shell command available on the system or python code can be executed. This also includes the execution of processes on a remote machine, although all necessary steps (e.g. connecting to the remote via SSH) need to be defined by the user. In general, such behavior as described in section 3.1 is possible, but it is not a built-in feature of doit. Also, doit does not intend to provide the compute environment. Therefore, while in general the composition of workflows (see section 3.6) is easily possible via python imports, this only works for a single environment. The status of the execution can be monitored via the console. Here, doit will skip the execution of processes which are up-to-date and would produce the same result of a previous execution. To determine the correct order in which processes should be executed, doit also creates a directed acyclic graph (DAG) which could be used to visualize dependencies between processes using “doit-graph” (https://github.com/pydoit/doit-graph), an extension to doit. For each run (specific instance of the workflow), doit will save the results of each process in a database. However, the tool does not provide control over what is stored in the database. On the one hand, doit allows to pass results of one process as input to another process directly, without creating intermediate files, so it is not purely file-based. On the other hand, the interface for non-file based inputs does not define the data type.
5.4 Guix Workflow Language
The Guix Workflow Language (GWL) [20] is an extension to the open source package manager GNU Guix [41]. GWL leverages several features from Guix, chief among these is the compute environment management. Like Guix, GWL only supports GNU/Linux systems.
GWL can automatically construct an execution graph from the workflow process input/output dependencies but also allows a manual specification. Support for HPC schedulers via DRMAA1 is also available.
GWL doesn’t provide a graphical user interface, interactions are carried out using a command-line interface in a text terminal. Monitoring is also only available in the form of simple terminal output.
There is support to generate a GraphViz (see e.g. https://graphviz.org) description of the workflow, which allows basic visualization of a workflow. Although not conveniently exposed2, GWL has a noteworthy unique feature inherited from Guix: precise software provenance tracking. Guix contains complete build instructions for every package (including their history through git), which enables accounting of source code and the build process, like for example compile options, of all tools used in the workflow. Integrity of this information is ensured through cryptographic hash functions. This information can be used to construct data provenance graphs with a high level of integrity (basically all userspace code of the compute environment can be accounted for [42]).
GWL uses Guix to setup compute environments for workflow processes. Each process is executed in an isolated3 compute environment in which only specified software packages are available. This approach minimizes (accidential) side-effects from system software packages and improves workflow reproducibility. Interoperability also benefits from this approach, since a Guix installation is the only requirement to execute a workflow on another machine. As Guix provides build instructions for all software packages, it should be easily possible to recreate compute environments in the future, even if the originally compiled binaries have been deprecated in the meanwhile (see [43] for a discussion about long-term reproducibility).
Composition of workflows is possible, workflows can be imported into other workflows. Composition happens either by extracting individual processes (repurposing them in a new workflow) or by appending new processes onto the existing workflow processes.
GWL relies exclusively on files as interface to workflow processes. There’s no support to exchange data on other channels, as workflow processes are executed in isolated environments.
Like other WfMSs, GWL caches the result of a workflow process using the hash of its input data. If a cached result for the input hash value exists, the workflow processes execution is skipped.
GWL is written in the Scheme [44] implementation GNU Guile [45], but in addition to Scheme, workflows can also be defined in wisp [46], a variant of Scheme with significant whitespace4. wisp syntax thus resembles Python, which is expected to flatten the learning curve a bit for scientific audience. However, error messages are very hard to read without any background in Scheme. On first use, GWL will be very difficult in general. This problem is acknowledged by the GWL authors and might be subject to improvements in the future.
As both wisp and Scheme code is almost free of syntactic noise in general, workflows are almost self-describing and easily human-readable.
In summary, GWL provides a very interesting and sound set of features especially for reproducibility and interoperability. These features come at the cost of a Guix installation, which requires administrator privileges. The workflow language is concise and expressive, but error messages are hard to read. At the current stage, GWL can only be recommended to experienced scheme programmers or to specialists with high requirements on software reproducibility and integrity.
5.5 Nextflow and Snakemake
With Nextflow [21] and Snakemake [10], the workflow is defined using a DSL which is an extension to a generic programming language (Groovy for Nextflow and Python for Snakemake). Moreover, Nextflow and Snakemake also allow to use the underlying programming language to generate metadata programmatically. Thus, authoring scientific workflows with Nextflow or Snakemake is very easy.
The process to be executed is usually a shell command or an external script. The integration with various scripting languages is an import feature of Snakemake as well as Nextflow, which encourages readable modular code for downstream plotting and summary tasks. Also boilerplate code for command line interfaces (CLIs) in external scripts can be avoided. Another feature of Snakemake is the integration of Jupyter notebooks, which can be used to interactively develop components of the workflow.
Both tools implement a CLI to manage and run workflows. By default, the status of the execution is monitored via the console. With Nextflow, it is possible to monitor the status of the execution via a weblog. Snakemake supports an external server to monitor the progress of submitted workflows.
With regard to the execution of the workflow (section 3.1), the user can easily run the workflow on the local machine and the submission via a resource manager (e.g. Slurm, Torque) is integrated. Therefore, individual process resources can be easily defined with these tools if the workflow is submitted on a system where a resource manager is installed, i.e. on a traditional HPC cluster system. Despite this, only level two of the defined criteria is met for Nextflow, since the execution of the workflow on a remote machine and the accompanied transfer of data is not handled by the tool. For Snakemake, if the CLI option “default-remote-provider” is used, all input and output files are automatically down- and uploaded to the defined remote storage, such that no workflow modification is necessary.
The requirement up-to-dateness is handled differently by Nextflow and Snakemake. By default, Nextflow recomputes the complete workflow, but with a single command-line option existing results are retrieved from the cache and linked such that a re-execution is not required. In this case, Nextflow allows storing multiple instances of the same workflow upon variation of a configuration parameter. Snakemake will behave like a build tool in this context and skip the re-execution of processes whose targets already exist and update any process whose dependencies have changed.
A strong point of Nextflow and Snakemake is the integration of the conda package management system and container technologies like Docker. For example, the compute environment can be defined for each process based on a conda environment specification file or a certain Docker image. Upon execution of the workflow, the specified compute environment is re-instantiated automatically by the WfMS, making it very easy to reproduce results of or built upon existing workflows. Furthermore, since the tool is able to deploy the software stack on a per process basis, the composition of hierarchical workflows as outlined in section 3.6 is possible.
Similar to doit, both tools do not provide a GUI to graphically create and modify workflows. However, a visualization of the workflow, i.e. a dependency graph of the processes, can be exported. Moreover, it is possible to export extensive reports detailing the provenance of the generated data.
Nextflow and Snakemake can also be regarded as file-based workflow management systems. Therefore, interface formats, i.e. class structures or types of the parameters passed from one process to the subsequent one, are not clearly defined.
5.6 Evaluation matrix
The evaluation of the WfMSs provided in section 5 in terms of the requirements described in section 3 on the example of the workflow outlined in section 4 yields the evaluation matrix depicted in table 1.
Table 1: Evaluation of the considered workflow management systems.
| Requirement | Workflow Management System | |||||
| AiiDA | CWL | doit | GWL | Nextflow | Snakemake | |
| Job scheduling system |  |
 |
 |
 |
 |
 |
| Monitoring |  |
 |
 |
 |
 |
 |
| Graphical user interface |  |
 |
 |
 |
 |
 |
| Provenance |  |
 |
 |
 |
 |
 |
| Compute environment |  |
 |
 |
 |
 |
 |
| Composition |  |
 |
 |
 |
 |
 |
| Process interfaces |  |
 |
 |
 |
 |
 |
| Up-to-dateness | L | R | U | U | L | U |
| Ease of first use |  |
 |
 |
 |
 |
 |
| Manually editable |  |
 |
 |
 |
 |
 |
6 Summary
In this work, six different WfMSs (AiiDA, CWL, doit, GWL, Nextflow and Snakemake) are studied. Their performance is evaluated based on a set of requirements derived from three typical user stories in the field of computational science and engineering. On the one hand, the user stories are focusing on facilitating the development process, and on the other hand on the possibility of reusing and reproducing results obtained using research software. The choice for one WfMS or the other is strongly subjective and depends on the particular application and the preferences of its developers. The overview given in table 1 together with the assessments in section 5 may only serve as a basis for an individual decision making.
For researchers that want to start using a WfMS, an important factor is how easy it is to get a first workflow running. We note again that the evaluation criteria of the requirement for the ease of first use are difficult to measure objectively and refer to section 3.9 for what is considered easy to use in this work. For projects that are written in Python, a natural choice may be doit, which operates in Python and is easy to use for anyone familiar with the language. Another benefit of this system is that one can use Python functions as processes, making it possible to easily transfer data from one process to the other via memory without the need to write and read to disk. In order to make a workflow portable, developers have to provide additional resources that allow users to prepare their environment such that all software dependencies are met, prior to the workflow execution.
To create portable workflows more easily, convenient tools are Nextflow or Snakemake, where one can specify the compute environment in terms of a conda environment file or a container image on a per-process basis. They require to learn a new domain-specific language, however, our assessment is that it is easy to get started as only little syntax has to be learned in order to get a first workflow running.
The strengths of AiiDA are the native support for distributing the workload on different (registered) machines, the comprehensive provenance tracking, and also the possibility to transfer data among processes without the creation of intermediate files.
CWL has the benefit of being a language standard rather than a specific tool maintained by a dedicated group of developers. This has led to a variety of tooling developed by the community as e.g. editors for visualizing and modifying workflows with a GUI. Moreover, the workflow description states the version of the standard in which it is written, such that any interpreter supporting this standard should execute it properly, which reduces the problem of version pinning on the level of the workflow interpreter.
Especially for larger workflows composed of processes that are still under development, and are thus changing over time, it may be useful to rely on tools that allow to define the process interfaces by means of strongly-typed arguments. This can help to detect errors early on, e.g. by static type checkers. CWL and AiiDA support the definition of strongly-typed process interfaces. The rich set of options and features of these tools make them more difficult to learn, but at the same time expose a large number of possibilities.
7 Outlook
This overview is not meant to be static, but we plan to continue the documentation online in the git repository [8] that contains the implementation of the exemplary workflow. This allows us to take into consideration other WfMSs in the future, and to extend the documentation accordingly. In particular, we would like to make the repository a community effort allowing others to contribute either by modifications of the existing tools or adding new WfMSs. All of our workflow implementations are continuously and automatically tested using GitHub Actions https://github.com/BAMresearch/NFDI4IngScientificWorkflowRequirements/actions, which may act as an additional source of documentation on how to launch the workflows.
One of the challenges that we have identified is the use of container technology in the HPC environment. In most cases, the way users should interact with such a system is through a module system provided by the system administrators. The module system allows to control the software environment (versioning, compilers) in a precise manner, but the user is limited to the provided software stack. For specific applications, self-written code can be compiled using the available development environment and subsequently run on the system, which is currently the state of the art in using HPC systems. However, this breaks the portability of the workflow.
Container technology, employing the “build once and run anywhere” concept, seems to be a promising solution to this problem. Ideally, one would like to be able to run the container application on the HPC system, just as any other MPI-distributed application. Unfortunately, there are a number of problems entailed with this approach.
When building the container, great care must be taken with regard to the MPI configuration, such that it can be run successfully across several nodes. Another issue is the configuration of Infiniband drivers. The container has to be build according to the specifics of the HPC system that is targeted for execution. From the perspective of the user, this entails a large difficulty, and we think that further work needs to be done to find solutions which enable non-experts in container technology to execute containerized applications successfully in an HPC environment.
Furthermore, challenges related to the joint development of workflows became apparent. In this regard, strongly-typed interfaces are required in order to minimize errors and transparently and clearly communicate the metadata (inputs, outputs) associated with a process in the workflow. This is recommended both for single parameters, but it would be also great to extend that idea to files - not only defining the file type which is already possible within CWL - but potentially allowing a type checking of the complete data structure within the file. However, based on our experience with the selected tools, these interfaces and their benefits come at the cost of some form of plugin or wrapper around the software that is to be executed, thus possibly limiting the functionality of the wrapped tool. This means there is a trade-off between easy authoring of the workflow definition (e.g. easily executing any shell command) and implementation overhead for the sake of well-defined interfaces.
Another aspect is how the workflow logic can be communicated efficiently. Although all tools allow to generate a graph of the workflow, the dependencies between processes can only be visualized for an executable implementation of the workflow, which most likely does not exist in early stages of the project where it is needed the most.
An important aspect is the documentation of the workflow results and how they have been obtained. Most tools offer an option to export the data provenance graph, however it would be great to define a general standard supported by all tools as e.g. provided by CWLProv [47].
A further direction of future research may also be a better measure for the ease of (first) use. As stated in section 3.9 this is rather subjective and depends on the experience and skills of the user. One could possibly treat this requirement statistically by carrying out a survey of the users of the respective tools.
Data availability:
Data can be found here: https://doi.org/10.5281/zenodo.7790634
Software availability:
Software can be found here: https://github.com/BAMresearch/NFDI4IngScientificWorkflowRequirements
Notes
- Distributed Resource Management Application API https://www.drmaa.org [^]
- GWL doesn’t provide a command to export provenance graphs in any way, instead Guix needs to be queried for build instruction, dependency graphs and similar provenance information of a workflows software packages [^]
- By default, lightweight isolation is setup by limiting the PATH environment variable to the compute environment. Stronger isolation via Linux containers is also optionally available. [^]
- GWL is not a workflow language in the strict sense. At its core, it is a Scheme library that defines functions and objects for workflow composition (like processes, inputs, outputs, etc.). It allows workflows to be defined in both Scheme and wisp. [^]
Financial disclosure
None reported.
Conflict of interest
The authors declare no potential conflict of interests.
Acknowledgements
The authors would like to thank the Federal Government and the Heads of Government of the Länder, as well as the Joint Science Conference (GWK), for their funding and support within the framework of the NFDI4Ing consortium. Funded by the German Research Foundation (DFG) - project number 442146713. Moreover, we would like to thank Sebastiaan P. Huber, Michael R. Crusoe, Eduardo Schettino, Ricardo Wurmus, Paolo Di Tommaso and Johannes Köster for their valuable remarks and comments on an earlier version of this article and the workflow implementations.
Roles and contributions
Philipp Diercks: Investigation; methodology; software; writing - original draft; writing - review and editing.
Dennis Gläser: Investigation; methodology; software; writing - original draft; writing - review and editing.
Ontje Lünsdorf: Investigation (supporting); software; writing - original draft (supporting).
Michael Selzer: Writing - review and editing (supporting).
Bernd Flemisch: Conceptualization (supporting); Funding acquisition; Project administration; Writing - review and editing.
Jörg F. Unger: Conceptualization (lead); Funding acquisition; Project administration; Writing -original draft (supporting); Writing - review and editing.
References
[1] M. D. Wilkinson, M. Dumontier, I. J. Aalbersberg, et al., “The FAIR guiding principles for scientific data management and stewardship,” Sci Data, vol. 3, no. 1, Mar. 2016, ISSN: 2052-4463. DOI: http://doi.org/10.1038/sdata.2016.1810.1038/sdata.2016.18. [Online]. Available: https://doi.org/10.1038/sdata.2016.18.
[2] B. Mons, E. Schultes, F. Liu, and A. Jacobsen, “The FAIR principles: First generation implementation choices and challenges,” Data Intellegence, vol. 2, no. 1-2, pp. 1–9, Jan. 2020, ISSN: 2641-435X. DOI: http://doi.org/10.1162/dint_e_00023. [Online]. Available: https://doi.org/10.1162/dint_e_00023.
[3] European Commission and Directorate-General for Research and Innovation, Realising the European open science cloud : first report and recommendations of the Commission high level expert group on the European open science cloud. Publications Office, 2016. DOI: http://doi.org/10.2777/940154.
[4] N. P. Chue Hong, D. S. Katz, M. Barker, et al., FAIR Principles for Research Software (FAIR4RS Principles), https://doi.org/10.15497/RDA00068, version 1.0, May 2022. DOI: http://doi.org/10.15497/RDA00068. [Online]. Available: https://doi.org/10.15497/RDA00068.
[5] C. Goble, S. Cohen-Boulakia, S. Soiland-Reyes, et al., “FAIR Computational Workflows,” Data Intelligence, vol. 2, no. 1-2, pp. 108–121, Jan. 2020, ISSN: 2641-435X. DOI: http://doi.org/10.1162/dint_a_00033. eprint: https://direct.mit.edu/dint/article-pdf/2/1-2/108/1893377/dint_a_00033.pdf. [Online]. Available: https://doi.org/10.1162/dint_a_00033.
[6] L. Wratten, A. Wilm, and J. Göke, “Reproducible, scalable, and shareable analysis pipelines with bioinformatics workflow managers,” Nat Methods, vol. 18, no. 10, pp. 1161–1168, Sep. 2021, ISSN: 1548-7091, 1548-7105. DOI: http://doi.org/10.1038/s41592-021-01254-9. [Online]. Available: https://doi.org/10.1038/s41592-021-01254-9.
[7] J. Schaarschmidt, J. Yuan, T. Strunk, et al., “Workflow engineering in materials design within the battery 2030+ project,” Advanced Energy Materials, vol. 12, no. 17, p. 2102638, 2022. DOI: https://doi.org/10.1002/aenm.202102638. eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/aenm.202102638. [Online]. Available: https://onlinelibrary.wiley.com/doi/abs/10.1002/aenm.202102638.
[8] P. Diercks, D. Gläser, J. F. Unger, and B. Flemisch, NFDI4Ing Scientific Workflow Requirements, version 0.0.1, https://github.com/BAMresearch/NFDI4IngScientificWorkflowRequirements, Jul. 2022.
[9] L. Griem, P. Zschumme, M. Laqua, et al., “Kadistudio: Fair modelling of scientific research processes,” Data Science Journal, vol. 21, no. 1, 2022. DOI: http://doi.org/10.5334/dsj-2022-016.
[10] F. Mölder, K. P. Jablonski, B. Letcher, et al., “Sustainable data analysis with snakemake,” F1000Res, vol. 10, p. 33, Apr. 2021, ISSN: 2046-1402. DOI: http://doi.org/10.12688/f1000research.29032.2. [Online]. Available: https://doi.org/10.12688/f1000research.29032.2.
[11] E. Afgan, D. Baker, B. Batut, et al., “The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update,” Nucleic Acids Research, vol. 46, no. W1, W537–W544, May 2018, ISSN: 0305-1048. DOI: http://doi.org/10.1093/nar/gky379. eprint: https://academic.oup.com/nar/article-pdf/46/W1/W537/25110642/gky379.pdf. [Online]. Available: https://doi.org/10.1093/nar/gky379.
[12] M. R. Berthold, N. Cebron, F. Dill, et al., “Knime: The konstanz information miner,” ser. Data Analysis, Machine Learning and Applications : Proceedings of the 31st Annual Conference of the Gesellschaft für Klassifikation e. V., Albert-Ludwigs-Universität Freiburg, March 7-9, 2007, New York: Springer, 2007.
[13] E. Deelman, K. Vahi, G. Juve, et al., “Pegasus, a workflow management system for science automation,” Future Generation Computer Systems, vol. 46, pp. 17–35, 2015, ISSN: 0167-739X. DOI: http://doi.org/10.1016/j.future.2014.10.008. [Online]. Available: https://www.sciencedirect.com/science/article/pii/S0167739X14002015.
[14] S. P. Huber, S. Zoupanos, M. Uhrin, et al., “AiiDA 1.0, a scalable computational infrastructure for automated reproducible workflows and data provenance,” Sci Data, vol. 7, no. 1, Sep. 2020, ISSN: 2052-4463. DOI: http://doi.org/10.1038/s41597-020-00638-4. [Online]. Available: https://doi.org/10.1038/s41597-020-00638-4.
[15] M. Uhrin, S. P. Huber, J. Yu, N. Marzari, and G. Pizzi, “Workflows in AiiDA: Engineering a high-throughput, event-based engine for robust and modular computational workflows,” Nato. Sc. S. Ss. Iii. C. S., vol. 187, p. 110086, Feb. 2021, ISSN: 0927-0256. DOI: http://doi.org/10.1016/j.commatsci.2020.110086. [Online]. Available: https://doi.org/10.1016/j.commatsci.2020.110086.
[16] E. N. Schettino, pydoit/doit: Task management & automation tool (python), https://doi.org/10.5281/zenodo.4892136, Jun. 2021. DOI: http://doi.org/10.5281/zenodo.4892136. [Online]. Available: https://doi.org/10.5281/zenodo.4892136.
[17] M. A. Salim, T. D. Uram, J. T. Childers, P. Balaprakash, V. Vishwanath, and M. E. Papka, Balsam: Automated scheduling and execution of dynamic, data-intensive hpc workflows, https://arxiv.org/abs/1909.08704, 2019. DOI: http://doi.org/10.48550/ARXIV.1909.08704. [Online]. Available: https://arxiv.org/abs/1909.08704.
[18] A. Jain, S. P. Ong, W. Chen, et al., “FireWorks: A dynamic workflow system designed for high-throughput applications,” Concurrency Computat.: Pract. Exper., vol. 27, no. 17, pp. 5037–5059, May 2015, ISSN: 1532-0626, 1532-0634. DOI: http://doi.org/10.1002/cpe.3505. [Online]. Available: https://doi.org/10.1002/cpe.3505.
[19] S. Lampa, M. Dahlö, J. Alvarsson, and O. Spjuth, “SciPipe: A workflow library for agile development of complex and dynamic bioinformatics pipelines,” GigaScience, vol. 8, no. 5, Apr. 2019, ISSN: 2047-217X. DOI: http://doi.org/10.1093/gigascience/giz044. eprint: https://academic.oup.com/gigascience/article-pdf/8/5/giz044/28538382/giz044.pdf. [Online]. Available: https://doi.org/10.1093/gigascience/giz044.
[20] R. Wurmus et al., GUIX Workflow Language, https://guixwl.org, version 0.5.0, Jul. 2022.
[21] P. Di Tommaso, M. Chatzou, E. W. Floden, P. P. Barja, E. Palumbo, and C. Notredame, “Nextflow enables reproducible computational workflows,” Nat Biotechnol, vol. 35, no. 4, pp. 316–319, Apr. 2017, ISSN: 1087-0156, 1546-1696. DOI: http://doi.org/10.1038/nbt.3820. [Online]. Available: https://doi.org/10.1038/nbt.3820.
[22] J. Köster and S. Rahmann, “Snakemake—a scalable bioinformatics workflow engine,” Method. Biochem. Anal., vol. 34, no. 20, pp. 3600–3600, May 2018, ISSN: 1367-4803, 1460-2059. DOI: http://doi.org/10.1093/bioinformatics/bty350. [Online]. Available: https://doi.org/10.1093/bioinformatics/bty350.
[23] S. P. Sadedin, B. Pope, and A. Oshlack, “Bpipe: A tool for running and managing bioinformatics pipelines,” Method. Biochem. Anal., vol. 28, no. 11, pp. 1525–1526, Apr. 2012, ISSN: 1460-2059, 1367-4803. DOI: http://doi.org/10.1093/bioinformatics/bts167. eprint: https://academic.oup.com/bioinformatics/article-pdf/28/11/1525/16905290/bts167.pdf. [Online]. Available: https://doi.org/10.1093/bioinformatics/bts167.
[24] P. Ewels, F. Krueger, M. Käller, and S. Andrews, “Cluster flow: A user-friendly bioinformatics workflow tool [version 2; referees: 3 approved].,” F1000Research, vol. 5, p. 2824, 2016. DOI: http://doi.org/10.12688/f1000research.10335.2. [Online]. Available: http://dx.doi.org/10.12688/f1000research.10335.2.
[25] I. Jimenez, M. Sevilla, N. Watkins, et al., “The popper convention: Making reproducible systems evaluation practical,” in 2017 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW), 2017, pp. 1561–1570. DOI: http://doi.org/10.1109/IPDPSW.2017.157.
[26] O. Ben-Kiki, C. Evans, and I. döt Net, Yaml ain’t markup language (yaml) version 1.2, Accessed: 2022-08-31, version 1.2, https://yaml.org/spec/1.2.2/, 2021.
[27] M. R. Crusoe, S. Abeln, A. Iosup, et al., “Methods included, Standardizing computational reuse and portability with the common workflow language,” Commun. ACM, vol. 65, no. 6, pp. 54–63, Jun. 2022, ISSN: 0001-0782, 1557-7317. DOI: http://doi.org/10.1145/3486897. [Online]. Available: https://doi.org/10.1145/3486897.
[28] K. Voss, G. V. D. Auwera, and J. Gentry, Full-stack genomics pipelining with gatk4 + wdl + cromwell [version 1; not peer reviewed], slides, https://f1000research.com/slides/6-1381, 2017. DOI: http://doi.org/10.7490/f1000research.1114634.1. [Online]. Available: https://f1000research.com/slides/6-1381.
[29] J. Vivian, A. A. Rao, F. A. Nothaft, et al., “Toil enables reproducible, open source, big biomedical data analyses,” Nat Biotechnol, vol. 35, no. 4, pp. 314–316, Apr. 2017, ISSN: 1087-0156, 1546-1696. DOI: http://doi.org/10.1038/nbt.3772. [Online]. Available: https://doi.org/10.1038/nbt.3772.
[30] S. Lee, J. Johnson, C. Vitzthum, K. Kırlı, B. H. Alver, and P. J. Park, “Tibanna: Software for scalable execution of portable pipelines on the cloud,” Method. Biochem. Anal., vol. 35, no. 21, pp. 4424–4426, May 2019, ISSN: 1367-4803, 1460-2059. DOI: http://doi.org/10.1093/bioinformatics/btz379. eprint: https://academic.oup.com/bioinformatics/article-pdf/35/21/4424/31617561/btz379.pdf. [Online]. Available: https://doi.org/10.1093/bioinformatics/btz379.
[31] C. Geuzaine and J.-F. Remacle, “Gmsh: A 3-d finite element mesh generator with built-in pre- and post-processing facilities, The gmsh paper,” Int. J. Numer. Meth. Engng., vol. 79, no. 11, pp. 1309–1331, May 2009, ISSN: 0029-5981. DOI: http://doi.org/10.1002/nme.2579. eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/nme.2579. [Online]. Available: https://doi.org/10.1002/nme.2579.
[32] M. Alnaes, J. Blechta, J. Hake, et al., “The FEniCS project version 1.5,” Archive of Numerical Software, vol. 3, 2015. DOI: http://doi.org/10.11588/ans.2015.100.20553.
[33] N. Schlömer, Meshio: Tools for mesh files, https://doi.org/10.5281/zenodo.6346837, version v5.3.4, Mar. 2022. DOI: 10.5281/zenodo.6346837. [Online]. Available: https://doi.org/10.5281/zenodo.6346837.
[34] W. Schroeder, K. Martin, B. Lorensen, and Kitware, The visualization toolkit : an objectoriented approach to 3D graphics, 4th ed. Kitware, 2006.
[35] J. Ahrens, B. Geveci, and C. Law, “ParaView: An end-user tool for large-data visualization,” in The Visualization Handbook, Elsevier, 2005.
[36] P. Williams and Contributors, The tectonic typesetting system, https://tectonic-typesetting.github.io/en-US/, Accessed: 2022-06-02, 2022.
[37] S. P. Huber, aiida-shell, version 0.2.0, https://github.com/sphuber/aiida-shell, Jun. 2022.
[38] P. Amstutz, M. R. Crusoe, N. Tijanić, et al., Common Workflow Language, v1.0, https://doi.org/10.6084/m9.figshare.3115156.v2, Jul. 2016. DOI: http://doi.org/10.6084/m9.figshare.3115156.v2.
[39] I. Colonnelli, B. Cantalupo, I. Merelli, and M. Aldinucci, “StreamFlow: cross-breeding cloud with HPC,” IEEE Transactions on Emerging Topics in Computing, vol. 9, no. 4, pp. 1723–1737, 2021. DOI: http://doi.org/10.1109/TETC.2020.3019202.
[40] A. B. Yoo, M. A. Jette, and M. Grondona, “Slurm: Simple linux utility for resource management,” in Job Scheduling Strategies for Parallel Processing, D. Feitelson, L. Rudolph, and U. Schwiegelshohn, Eds., Berlin, Heidelberg: Springer Berlin Heidelberg, 2003, pp. 44–60, ISBN: 978-3-540-39727-4.
[41] L. Courtès, “Functional package management with guix,” European Lisp Symposium, Jun. 2013. DOI: http://doi.org/10.48550/ARXIV.1305.4584. [Online]. Available: https://arxiv.org/abs/1305.4584.
[42] L. Courtès, “Building a secure software supply chain with GNU guix,” The Art, Science, and Engineering of Programming, vol. 7, no. 1, Jun. 2022, ISSN: 2473-7321. DOI: http://doi.org/10.22152/programming-journal.org/2023/7/1. [Online]. Available: https://doi.org/10.22152/programming-journal.org/2023/7/1.
[43] M. Akhlaghi, R. Infante-Sainz, B. F. Roukema, M. Khellat, D. Valls-Gabaud, and R. Baena-Galle, “Toward long-term and archivable reproducibility,” Computing in Science & Engineering, vol. 23, no. 3, pp. 82–91, May 2021, ISSN: 1521-9615, 1558-366X. DOI: http://doi.org/10.1109/mcse.2021.3072860. [Online]. Available: https://doi.org/10.1109/mcse.2021.3072860.
[44] M. Sperber, R. K. Dybvig, M. Flatt, A. Van Straaten, R. Findler, and J. Matthews, “Revised6 report on the algorithmic language scheme,” J. Funct. Program., vol. 19, no. S1, p. 1, Aug. 2009, ISSN: 0956-7968, 1469-7653. DOI: http://doi.org/10.1017/s0956796809990074. [Online]. Available: https://doi.org/10.1017/s0956796809990074.
[45] A. Wingo et al., GNU Guile, https://www.gnu.org/software/guile/, Feb. 2022.
[46] A. Babenhauserheide, Srfi 119: Wisp: Simpler indentation-sensitive scheme, https://srfi.schemers.org/srfi-119/, Jun. 2015.
[47] F. Z. Khan, S. Soiland-Reyes, R. O. Sinnott, A. Lonie, C. Goble, and M. R. Crusoe, “Sharing interoperable workflow provenance: A review of best practices and their practical application in cwlprov,” GigaScience, vol. 8, no. 11, Nov. 2019, ISSN: 2047-217X. DOI: http://doi.org/10.1093/gigascience/giz095. [Online]. Available: https://doi.org/10.1093/gigascience/giz095.